9:25a – 11:50p
Probe Making
- very poor probe incorporation. Maybe the lower concentration of Maxima?
- Try to run RT again tomorrow to compare.
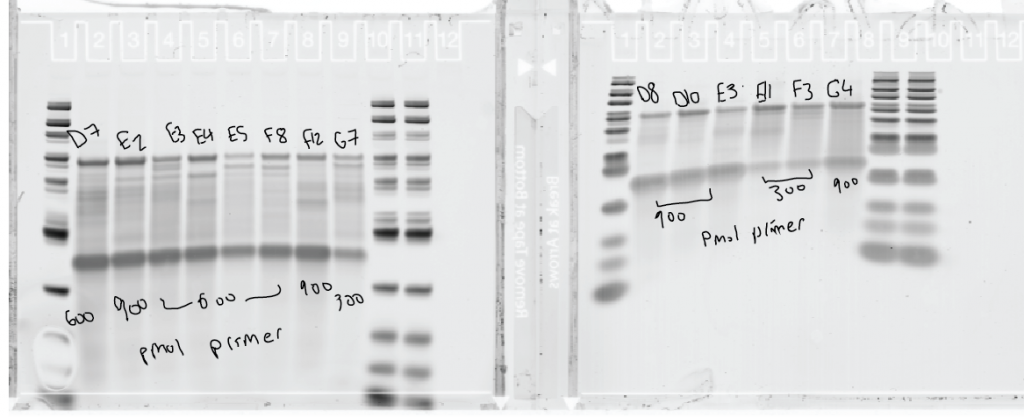
- attempted DCC of probes to remove excess primer using short probes 1-4. Extremely low yield (20 ng/uL instead of 500-600).
Deep sequencing
- Ran samples on MiSeq
- Csv parameter file in email
- needed to update MiSeq software for V3 kit.
- Ran 12 pM scale reaction
- Sequencer seems to be running smoothly as of this evening.
Ph Project
- put antibodies aside for Ajaz.
STORM analysis
- E01 data – 647 conventionals are useless. Staining looks pretty weak on deeper investigation, not sure I trust this probe. Should remake it. Examination of molecule switching looks sub-par as well — not a great STORM run either. Replaced E01 on microscope for investigation. Staining does not look good.
- E10 beads not analyzed — set to running on Cajal
- E08 data and beads not yet analyzed — set to running on Cajal
- qued up to many files, Cajal streaming data slowly.
- Canceled transfer of G1 data. Need to resume tomorrow (too much traffic to ProBox)
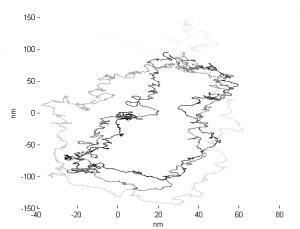
- Ran D12 data through analysis
- Periodic drift pattern on STORM4
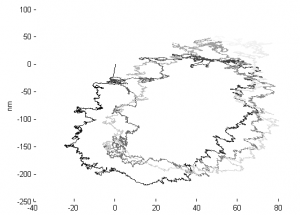
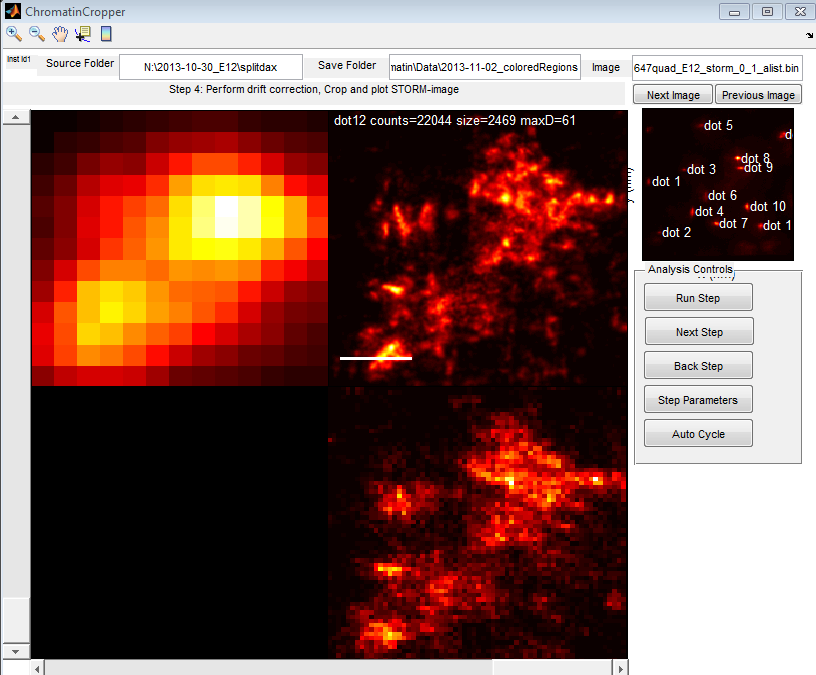
- started E12 analysis (lots of dots)
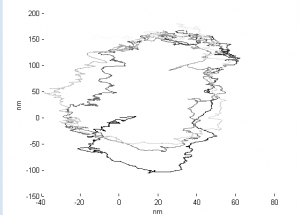
STORM imaging
Cell Culture
- Kc167 cells, growing alright but non-adherent
- Kc cells grow better in SFX media than Schneider’s. Ordered SFX.
- Requested new Kc cells. Might as well start with a clean line from the DGRC
- mix up ConA solution at 0.2 mg/mL (100x). 2 ug/mL is supposed to aggregate RBCs.
- UV sterilize ConA, then coat 6-well plates with ConA and add cells. See if we get any better adherence.