8:30a – 7:30p,
8:30p – 11:30p (remotely)
Goals
- Write to Alvaro (done)
- Reply to Zenklusen (done)
- Continue analysis of MiSeq Results (in process)
- write to Jeff to get gene list (done)
- repeat RT reaction with more enzyme (done)
- Work on modeling figures for Ph Project (worked out sketch with Ajaz)
- start imaging cells for Ph project on STORM2 (cells failed)
- Finish editing Banff Workshop report and send around for final review prior to submission. (tomorrow?)
- Double check requirements for report submission. (tomorrow?)
- Flip Fly Stocks! (tomorrow?)
STORM analysis
- Some G-630 regions don’t fit in the space of the viewer, area’s are artificially smaller.
- Mildly concerned that some regions elongated in y by 3D optics get incorrectly parsed by DaoSTORM.
Ph Project
- meeting with Ajaz
- see post for figure changes etc to work on
- no cells found on Ph-slide
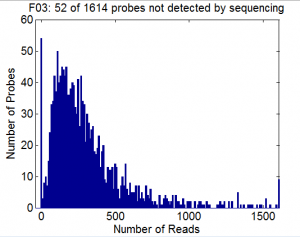
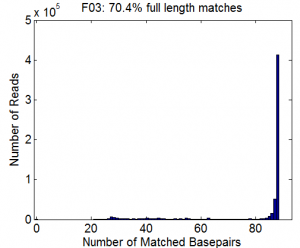
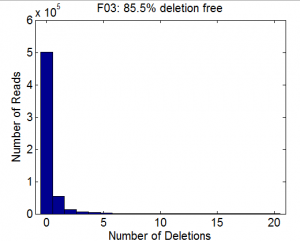
Library Analysis by Deep Sequencing
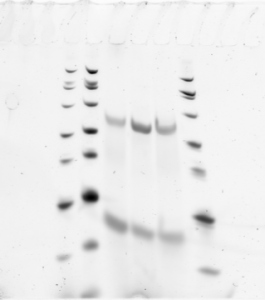
Probe Making tests
- More maxima does not improve yield. RNA quality is everything.
- samples D08, D10, E03, (organized in reverse order on the gel. The double ladder is always loaded last).
Deep Sequencing Analysis and Coding
- Wrote function
MapSeqReads.mto analyze coverage and mutation rate for library and sublibraries. - Fully functionalized form with help file and array of optional inputs. Still needs some debugging probably
- target matching not working well at all for mouse libraries.