10:45a – 12:15a
Notebook
- backed up notebook
- updated wordpress
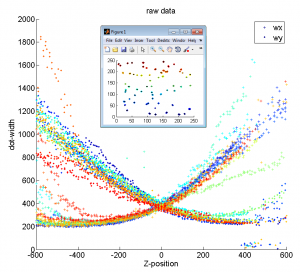
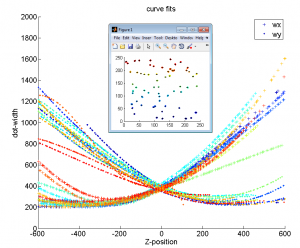
Z-calibration
- cross correlation doesn’t improve curve alignment visibly — curves are fundamentally a bit different
- no spatial structure evident
Ph Project
- reviewing simulation results
- saved as
simulationName_2.epsto file
Test new stains
- at higher concentration, probes work great. Will need to make more though, total yield here was actually quite poor then. Try splitting RT into smaller reactions.
- Beads at 1:1000 this time way too dense. Single beads look fine on glass, but on cells this time the beads are coated too dense to separate one from another.
- try again at high concentration to do the 750 labeling. Try D12-D12 750 + 647.
probe making
- spec’d concentrations of primers but otherwise failed to start PCRs
STORM analysis
- worked through black regions, added images of 4 black regions (10, 18, 180, and 425 kb) to presentation
- helping Guipeng get DaoSTORM working
- need to sync up matlab-storm versions, push to master run clean install. (could create dummy user on Tuck to test?)
Project 2
- Ran a variety of error analysis with Hao
- Computed error over time — confirmed no detectable / statistically significant increase.