9:00a – 12:15a
Goals
- Write to Ajaz about new plans
- Read in _oligo.txt files and analyze probe distributions
- STORM G1-G2
- Start analysis of F12-G01 and F12-G11 data
- continue work on STORMrender Manual
Literature
- Reading Kieffer-Kwon et al Cell 2013. link
- Really, insulators vary little across cell types? I don’t think this is true. Maybe it’s just lack of data at present.
- The fundamental question of how enhancers know to talk to the right promoters seems largely un-addressed. Do we have other hypotheses aside from 3D spatial proximity? 3D spatial proximity guided by compartment formation
- with all the talk about ‘looping out’ one would expect these HiC maps to have more check-board and less block-like structure…
Library 3 design
- 78,225 probes using non-stringent t80 x78 s78 parameters. Room for more genome regions.
- still need to add some black regions by parts
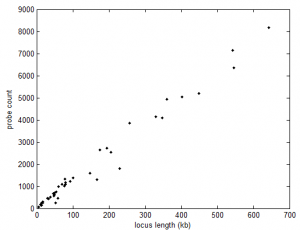
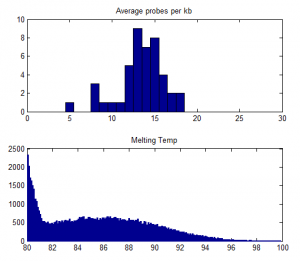
- some preliminary stats:
Probe making
- RT reactions, samples 1-8, 400 pmol primer with 10 uL of RNA solution. ~50% incorporation.
- D12, F11, F12, G1, G2, G3, G5, G6, P1 and P3
- F03, F04, F05, F06, G04
STORM analysis
- running dot fitting for F12-F11 data.
- serious issues with buffer / dye brightness. I also swear these looked better during acquisition on storm2.
matlab-storm
- updated STORMfinder to add an Options menu for controlling analysis parameters
Other
- updating professional webpage