9:20a -7:00p
Goals
- Analyze 2-color STORM data
- test out new image-writers for hal
Getting started
- Monet windows explorer froze and got killed. Requires restart.
- Finish up notes from yesterday, add gel images to notebook.
STORM
- attempted software update to do bead-averaging on-the-fly. Failed.
- STORM2 software several updates behind, frame objects have been substantially redefined.
- More consultation with Hazen, updates will require some substantial reconfiguring. Needs to wait until Colenso’s data collection is finished.
- github not yet set up on STORM2, let’s just try this manually for now.
Analyzing F11-F12 STORM data
- Starting with image _0_0, 647 channel having trouble being well resolved in STORM images. Not sure why?
- observation 1, 647 hardly visible in conventional image. I know this slide had good labeling, clear conventional dots. Not sure what’s up.
- trimming down the number of frames seems to improve the quality in some cases. I think some images exhausted the low level labeling in the first three or five thousand frames and spend the rest of the movie accumulating background
- background is legitimately quite high in theses movies.
coding matlab-storm
- working on building a demo-data set
- don’t really have impressive multi-color STORM images that I’m ready to share openly with the public.
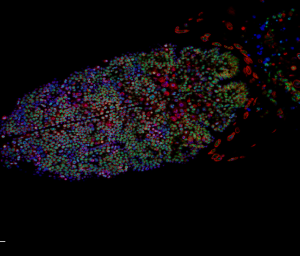
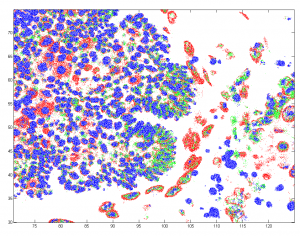
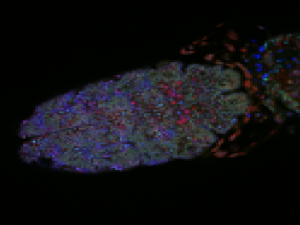
- building a demo data from down-sampled confocal data of Drosophila embyro — this should illustrate most of the points.
Literature
- Continue reading Kieffer-Kwon et al Cell 2013.
- still, a very nice data set. Best display of ChIA-PET data I’ve seen, the chip-seq tracks with the red lines linking peaks works well. The promoter enhancer culstering graphs and the complete array organized by complexity are also very nicely done.
- Types of PolII groups — extragenic – tethering promoters to distal enhancers; intragenic, connecting promoters to gene bodies — (don’t you think these are intronic enhancers? they are rather clear peaks from intronic sequence more than general spread across the gene body) + promoter-promoter and enhancer + enhancer.
- Though on closer examination I see that they see a clear, though subtle increase throughout the gene body as well of transcribed genes, so maybe this is what is referred to. In which case I think it is very likely enzyme accessibility.
- how much of this is open chromatin driven? Maybe open chromatin is the biological basis of selection?
- okay, also later on they do refer examples of an intronic enhancers. So you could call it 5 classes and be clearer, or not define enhancer contacts as necessarily ‘extragenic’.
- 90% of enhancers linked to a single promoter, less than 2% linked to more than 2, max = 7. (what do you expect by chance as a function of local density of open chromatin regions?)
- I wonder how many of these promoter promoter contacts are promoter proximal enhancer elements?