9:50a – 12:15a
Chromatin Project
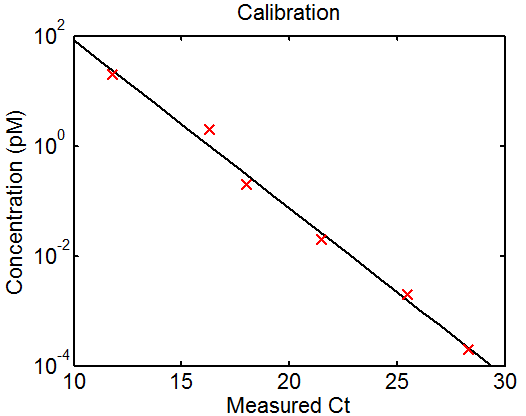
Deep sequencing
QPCR
- run QPCR
- quantify results from QPCR
C:\Users\Alistair\Documents\Research\Projects\Chromatin\Code\Scripts\qPCR_MiSeq2Prep_140212.m
MiSeq Prep
- sample concentrations / calculating dilutions:
- orig library
- raw 10 uL scale reaction: 25, 14, 25, nM
- dilutions 1:10 diluted: 1.98, 2.0, 2.0
- these are 2x listed concentrations since its a 10 uL not a 20uL reaction volume
- prep samples for MiSeq run
- Start MiSeq
Meetings
- discussion with Jane and Bogdan about chromatin data and models
- random walks are non-spherical: should look more at elipticity data
- try log-log plot with 1/3rd 1/2 and 1 lines.
- schedule STORM users meeting for Monday. Need to organize some lists.
- Need to measure max laser powers on STORM2 and STORM4 for Monday meeting
Cell staining
Cell prep
- removed cells from RNase treatment (9p-9a at 37C, hope this wasn’t too much for the delicate structures).
- SSC treatment. moved to formamide.
- Prehybed ~1hr at 47C.
- small hybe oven having issues (shot up to 58 when I removed the three large water chambers).
- Using large drying oven for better thermal stability
Probes
- G01-G02-G03-P1 A647
- F03 + F04-P1 A647
- G05-P1 + G06-P3 + S1-A647 + S2-A750
- G05-P3 + G06-P1 + S1-A647 + S2-A750
- F05-P1 + F06-P3 + S1-A647 + S2-A750
- F05-P6 + F06-P1 + S1-A647 + S2-A750
Secondary projects
Ph Project
Cell staining
- (7) S2 cells: G03P2-cy3 + F03P3 + F04P3 + F05P3 + S3-A750
- (8) Ph-WT 1 cells: G03P2-cy3 + F03P3 + F04P3 + F05P3 + S3-A750
- (9) Ph-WT 2 cells: D12-cy3
- all at 0.4 – 0.6 uL of 100 uM secondary + 1-2 uL of primary (each in case of multiples).
OligoPaint project
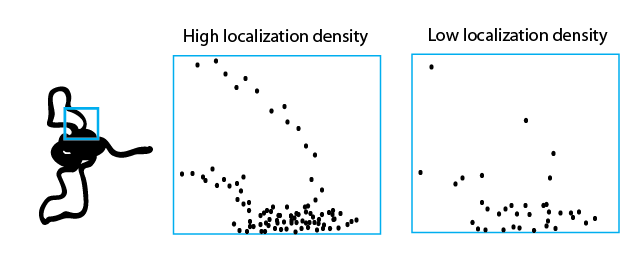
- need better explanation of STORM
- sent fig from XZ review to BB.
- Also this: