10:15a – 5:50p
Chromatin Project
Library 3 prep
- finish annotating new lib3 yellow green regions
- ran probe folding for yellow green regions
- Current probes (including 10 new black, 8 new blue): 91144
Deep Seq analysis
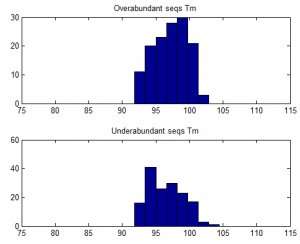
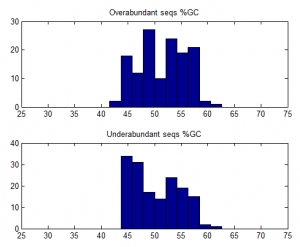
- Trying to understand why some sequences are consistently over-abundantly amplified.
- no effect of number of hairpins or number of dimers
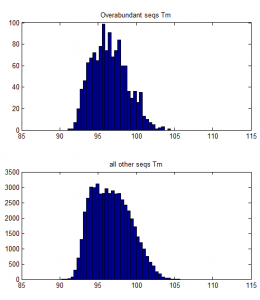
- weak affect of Tm / %GC. Higher TM more GC more a bit more frequently abundant on ave.
over-abundant after PCR vs all other seqs:
Analyzing data from Hao Lib
- see post
Data cleanup
- running splitdax on old bead data (this is the only way we can compress the bead movies).
- using D11 as test set.
Ph project
- spot checking of half a dozen or so of the S2 and WT Ph-STORM + FISH experiments don’t show obvious colocalization. The labeling is obviously sparser, (maybe 1/5 the number of clusters) probably just the brightest ones.