10:15a – 12:30a
Goals
- Data processing
- copy C05 and C06 data to hard-drives
- Launch analysis of C03-C06 data
- plate, fix, prep and hybe new cells
- Probe making
- RT reactions
- test gel
- probe purfication
- cell staining
- wash out probes
- image multi-color cells on STORM2
Chromatin Project
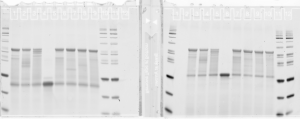
Probe making.
- finished making 16 new probes (2 failed, no template).
- gel looks pretty uniform for concentration
- F03 has weird series of bands — probably not good. Should remake. F05 failed completely (no template, I guess I knew this. Good negative control).
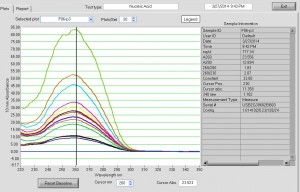
- attempted 500 pmol scale synthesis. Incorporation looks less good, should stay at 400 pmol per 10 uL RNA.
Cell staining
- fixed and prepped new cells for staining, including 1hr+ RNase A treatment at 37C.
- F03-p3 2 uL, F02-p1 7 uL
- F05 P3 1.5 uL, F06-P1 6 uL
- F06 P3 2.5 uL, F07-P1 6 uL
- Damn it I used the failed and questionable probes in the staining. Need to check these things!
- Why do I get better yield on the failed probes?
Imaging
C07 on STORM2
- 3 uL COT, 5 uL BME,
- looks reasonable
- Need to remember to take beadfields in the morning! Otherwise there will be lots of beads to manually parse out of ChromatinCropper.
C08 on STORM1
- 2 uL COT, 5 uL BME
- spots identifiable in all cells, but weak / small / low contrast.
- realigned back-optics (way off center)
- tweaked dual view to separate channels Cy5Cy3 3D. Alignment is off by 10s of pixels between channels.
- Need to remember to take beadfields in the morning! Otherwise there will be lots of beads to manually parse out of ChromatinCropper.
Coding / Research Organization
- established GitScripts repo on github, pushed scripts. Now these and their revision histories backed up somewhere. Yay!
- pulled latest version of script databases into GitScripts and committed and pushed up to Github (This takes a couple commands, including a move command to get new scripts to there appropriate folders. I bet I can write a matlab wrapper to do this in a single click.)
- added private protocols to GitScripts