9:45a – 11:50p
Goals
- Record bead fields.
- check isoforms differences and consequences for probes selected (do our probes still hit the most popular isoform of imr90s?)
Cell Staining
- washout stains for new multi-color samples. Need to wait until oven is free.
Imaging samples
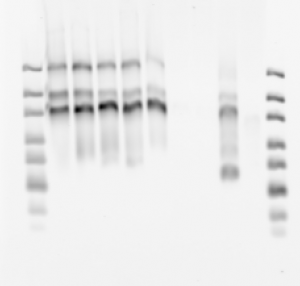
- F03-p3 2 uL, F02-p1 7 uL — F02-P1-A647 actually works (first time!) should image this. F03-P3-cy7 is detectable but very dim. I have gotten much brighter A750 stains before. Background is also high.
- F05 P3 1.5 uL, F06-P1 6 uL — F05 probe synthesis failed. F06-P1 looks strong and clear at thic concentration.
- F06 P3 2.5 uL, F07-P1 6 uL — F06-P3-cy7 failed ? High background. F07-P1-A647 looks fine. Not clear why F06 works with P1 and not with P3.
New Stains
- F3-6, F1-7 6 uL, 6 uL
- G6-6, G8-7
- G8-6, G6-7
- F7-6, F6-7
- F6-6, F7-7
- G2-6, G3-7 1.5 uL, 1.5 uL (~1100 ng/uL)
New Probes
- E01-E08
- chr3R:19929199-20187132__YELLOW_250kb_silent_flanks
- chrX:1907754-2083414__YELLOW_175kb_silent_flanks
- chr3R:19929199-20029199__YELLOW_250pt1_100kb
- chrX:1907754-1977754__YELLOW_175pt1_70kb
- chrX:1977754-2047754__YELLOW_175pt2_70kb
- chr2R:19726615-19787585__YELLOW_D12a
- chr2R:19809874-19888410__YELLOW_D12b
- chr2R:19906491-19976552__YELLOW_D12c
- Neg control, E01fwd, E07rev
- ran PCRs
- started T7 reactions
Not sure why E06 and E07 failed, and E08 looks weird. Maybe press on with just E01-E05.
Research Organization
- moved ChromatinCropper into matlab-functions
- moved commonly used mature functions out of Beta into appropriate matlab-functions folder (e.g. normhistall is now in Plotting).
- moved storm-analysis and storm-control into Software.
- moved Scratch into Software
- moved GeneralSTORM scope images onto Data and archived code into Archive2014 on D drive