9:45 am – 1:30 am
Chromatin Modeling continued
Checking my simulation results from the polymer models I was running last night.
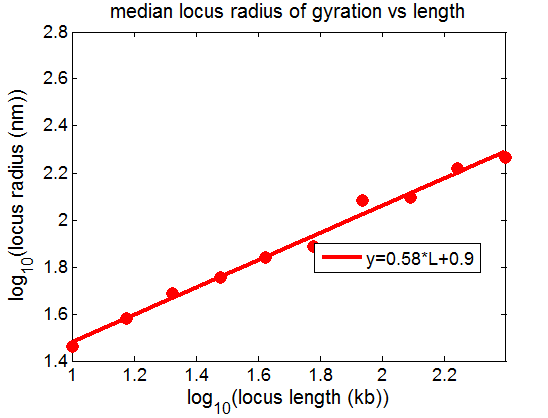
Simulation Results
BBv1 no attractive potential does give expected scaling:
Adding an attractive potential does increase clustering. Has more effect on large polymers.
Notes from meeting with Max
- Combine repulsive and attractive forces into a single potential.
- use modified square well
- Need to learn better use of python classes and namespaces. See Python class doc: https://docs.python.org/2/tutorial/classes.html
More coding
- Fixed up openmm-polymer code
- implemented extension script to import and build off the openmmlib (I called my function lib
openmmlibExt). - with some difficulty (stupidity), finally implemented node specific interactions
- simulating free polymers,
- simulating uniformly sticky polymers
- simulating periodically stick polymers
Primer ordering
- Lib4 primers still had the same bug as the Lib3 primers. Now when back and editted old scripts to fix this bug (will be logged in GitScripts).
- bug was in the export list of T7 rev primers, the sequences of the Fwd primers was used and not the rev primers.
- wrote new functions to index 96 well plates and to convert fasta files to csv tables for ordering primers.
- validated primers in quick test.
- remembered (belatedly) to add T7 to project 2 L5 primers and strip off extra Gs
- sent primer orders to Alec.
Staining
- new cell stains: C09 C10 and C11 (tiny blue and black regions)