November 2025 M T W T F S S « Aug 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 Categories
- AP patterning (13)
- Blog (1)
- Chromatin (88)
- Conference Notes (72)
- Fly Work (54)
- General STORM (25)
- Genomics (134)
- Journal Club (22)
- Lab Meeting (66)
- Microscopy (79)
- Notes (1)
- probe and plasmid building (58)
- Project Meeting (3)
- Protocols (13)
- Research Planning (74)
- Seminars (21)
- Shadow Enhancers (59)
- snail patterning (40)
- Software Development (5)
- Summaries (1,412)
- Teaching (9)
- Transcription Modeling (40)
- Uncategorized (10)
- Web development (19)
Links
Tags
analysis cell culture cell labeling chromatin cloning coding communication confocal data analysis embryo collection embryo labeling figures fly work genomics hb image analysis image processing images in situs Library2 literature making antibodies matlab-storm meetings modeling MP12 mRNA counting Ph planning presentation probe making project 2 project2 result results sectioning section staining shadow enhancers sna snail staining STORM STORM analysis troubleshooting writing-
GitHub Projects
Protected: Lab Meeting 05/29/15
Posted in Lab Meeting
Comments Off on Protected: Lab Meeting 05/29/15
Thursday 05/28/15
9:30 am – 9:45 pm
Manuscript Revisions
Discussions with Jeff and Bogdan
- move pairing discussion to end of volume scaling paragraph.
- (now addressed in version 8-5).
- show only volume not rg in extended data on higher order effects
- (BB will address in revised figure)
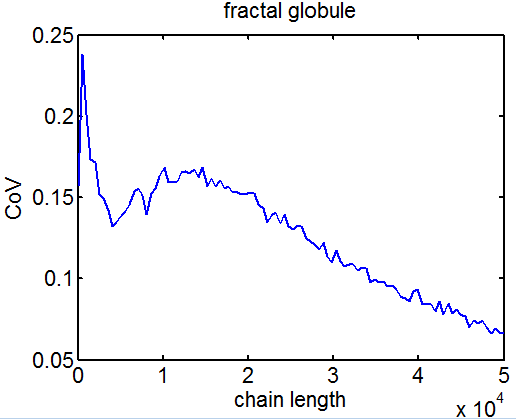
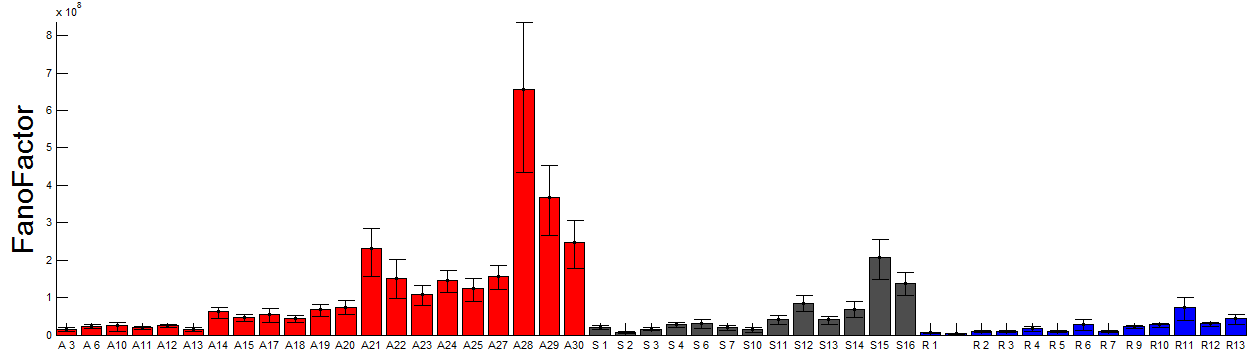
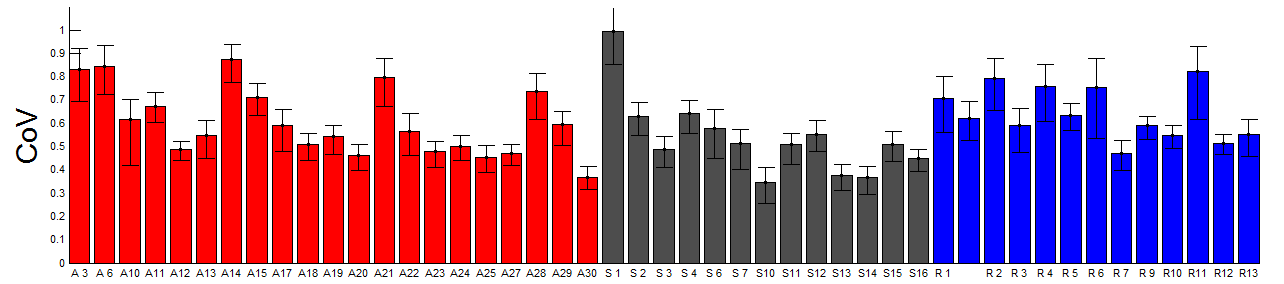
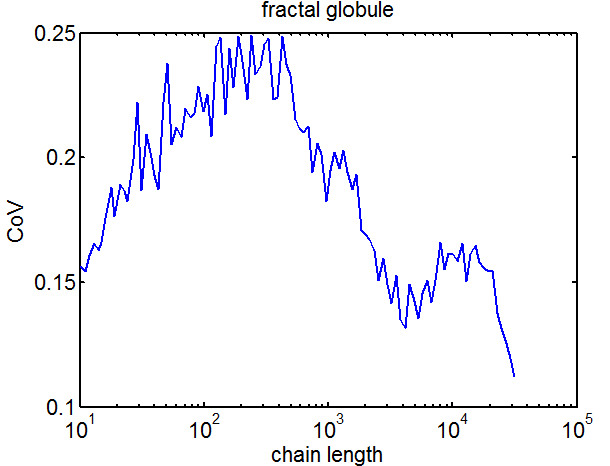
- add analysis of CoV to ED figure 4
Communications with Mirny team
- LE model does not have 0.3 rg scaling
- probably leave off for now?
writing
- added comment on comparison of differences of silent and repressed with HiC
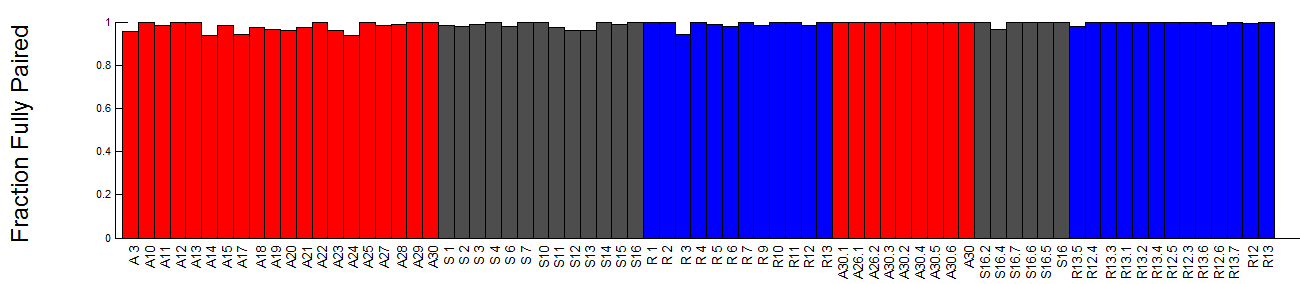
- revised discussion of pairing.
Figure modifications
- encorporated CoV analysis into ED figs 3 and 4.
Reorganizing
- reorganized code for fig 1 — all from allData.matb
- still need to track down some missing vlists
- identified 8 domains in our most recent list of domains that are still not included in allData. Includes a number of internal domains. Annotated these in excel. (NewMasterTable_1ColorData)
- need a similar structure for the 2 color data for Fig 2 and 3, can do tomorrow
- should convert the model simulations results to a matb files as well for easy access for BB
Conference Prep
- booked hotel for SDB
- booked flights for ASBMB
- still need to book flights for SDB. Kayak recommends wait.
Posted in Summaries
Comments Off on Thursday 05/28/15
Wednesday 05/27/15
9:30 am – 12:35 am
Organizing
- agree to review for JTB (will do next week after submission)
- troubleshooting STORMfinder on Mac
Analyzing cell-to-cell variation
Identifying split loci
Revising manuscript
- new figures:
- pairing analysis
- cell-cell variation
- higher order effects
- revised main text
- sent revised version to XZ, ~12:30 am
Posted in Summaries
Comments Off on Wednesday 05/27/15
Tuesday 05/26/15
8:30 am – 12:30 am
Revising chromatin manuscript.
- Figures all on version 10
- revised all figures as discussed except new E8
- embedded all figures in 8-2
- main text on version 8-2.
To do tomorrow
- finish responding to XZ comments in methods on polymer simulations
- read and respond to JM comments
- analysis split-spots / lose pairing (at all scales)
- write associated long and short form discussion
- analysis of deviation from line for all domains
- analysis of cell-cell variation for all domains and subdomains (both variance/mean, std/mean)
- incorporate all heterogeniety analysis into text
- rewrite correlation of TADs and epigenetic domains in intro
- rewrite section dealing with active domains following today’s discussion.
Posted in Summaries
Comments Off on Tuesday 05/26/15
Monday 05/25/15
10:00 am – 5:10 pm, 8:00 pm – 11:45 pm
Parallelizing image analysis
issues with parfor loops
- much faster
- easily exploading memory
- observation: launch and shutdown of parpool is not fast — takes several minutes even to spin down.
- Found issue: using matrices xDim x yDim x numGenes for indexing is is too memory intensive for 1001 genes and parallel pool of 20 cells.
- no global variables allowed inside parfor loops
success
- now running efficiently through all data.
- uses up to 20 GB memory and 65% CPU
- runs rather slower on 1000 gene data than 140 gene data: loop over genes are slowing things down a bit.
- slow step is still the intial warp and deconvolution — this can run pretty efficiently overnight though.
- Morgan may reboot itself before my code finishes though…
Chromatin paper
- edits back from XZ. Make changes for tomorrow.
Posted in Summaries
Comments Off on Monday 05/25/15
Friday 05/22/15
8:00 am – 5:00 pm, 8:00 pm – 9:45 pm
Goals
- revise domain names to match new conventions
- write 100 word summary for paper.
Meetings
- lab meeting
- discuss microscopes
- group C presentations
- microscope decisions
- buy new Andor 897 ultra for STORM5
- buy new 750 laser (MPB if they can promise time) for STORM5
- buy new 1-2W 561 laser (MPB) for STORM3
- I will help CX install new camera and quadview and set up 3D imaging on STORM2.
MERFISH coding
- working on converting new approach into a discrete function based pipeline of code
- plan to wrap these functions into a GUI as well.
- Additional approach: using blanks formally in a machine learning context to discriminate real spots from background
- we have examples of what we know for certain background looks like, and examples of stuff we think is signal but may include background.
- we can set a false positive tolerance (number of background counted) and chose the discriminator that maximizes detection while meeting this required false positive.
- current approach does completely independent cuts on total brightness and brightness ratio (two highly correlated quantities). Could compute the proper 2D (or ND) seperator between
Manuscript
To do:
- need to address JM’s outstanding comments
- got feedback from BB — some recommended reference changes. Need to add
- go feedback from GF and MI — some recommended reference additions. More with LM’s comments.
Computer observations
- Tuck and Cajal very slow after logging into terminal
- suspect windows / dropbox syncing configuration is slowing things down (?)
- Cajal when run at terminal just seems painful. Graphics card communication issue?
Posted in Summaries
Comments Off on Friday 05/22/15
Thursday 05/21/15
11:00 am – 12:00 am
(9am-11am lab volleyball practice)
Chromatin paper
- discuss revisions with Xiaowei
- working on critical revisions prior to sending to collaborators
- sent draft to CW and LM and team
MERFISH
- update planning meeting with XZ
- discussion with team
- should have a new go at addressing isoform variation.
Posted in Summaries
Comments Off on Thursday 05/21/15
Wednesday 05/20/15
9:00 am – 9:50 pm
MERFISH
Playing with deconvolution of Laplacian processing
- see notes
- summary: very promising in speed, accuracy, simplicity, and aesthetics.
- ran new pipeline on 141004 data (saved in folder 150520)
- rerunning with more stringent selection against background (in folder 150520b). 50 cells ~1 hour run time (largely in saving data).
- starting pipeline5 script — combine read & warp dax files with rest of analysis. Convert into functions.
For presentation
- Register for EMBO meeting on nuclear structure and submit abstract
- highlight differences in structure in summary
- “compact and diffuse domains, branched and linear domains, domains that are highly entangled with one-another and domains which are strictly segregated”
- highlight implications
- short range contacts in Active regions in cultured cells
- long range interactions in Polycomb domains
- hox genes more tightly packed than other genes because of colinearity. (better repressed)?
Posted in Summaries
Comments Off on Wednesday 05/20/15
Protected: MERFISH image processing 05/20/15
Posted in Genomics
Comments Off on Protected: MERFISH image processing 05/20/15
Nuclear Structure and Dynamics 2015 Abstract
Nuclear Structure and Dynamics
http://events.embo.org/15-nucleus/
Super-resolution Imaging of Chromatin Nanostructure Links Epigenetic State and 3D Genome Packaging
Metazoan genomes are packaged at multiple scales, and regulation of this spatial organization may play an important role in development and cell fate selection. Unfortunately, current methods provide little information about the 3D organization of the genome at the length scale of genes (kilobases) and regulatory domains (hundreds of kilobases) in single cells. I will present a new super-resolution imaging approach to study the structural organization of the genome at the kilobase to megabase scale in individual cells at 20 nm resolution. These domains largely occupy diffraction limited volumes and thus their structures cannot be resolved with conventional imaging approaches. From thousands of images of epigenetic domains across the Drosophila genome, we have discovered, within a single cell type, a substantial diversity of structural patterns: compact and diffuse domains, branched and linear domains, domains that are highly entangled with one-another and domains which are strictly segregated. These different structural features are closely correlated to certain differences in the epigenetic state of the chromatin. I will highlight the organization of Polycomb bound domains, which exhibit a surprising, entangled structure and length-dependent compaction. Computational models suggest this organization could contribute to the repressive nature of the domains. Preliminary comparisons between developing tissues suggests differential regulation of chromatin structure is associated with developmental fate selection. Together, our observations reveal rich structural diversity in chromatin packaging, suggest some potential regulatory factors, and highlight the importance of understanding structural organization for the study of gene regulation.
Posted in Summaries
Comments Off on Nuclear Structure and Dynamics 2015 Abstract