November 2025 M T W T F S S « Aug 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 Categories
- AP patterning (13)
- Blog (1)
- Chromatin (88)
- Conference Notes (72)
- Fly Work (54)
- General STORM (25)
- Genomics (134)
- Journal Club (22)
- Lab Meeting (66)
- Microscopy (79)
- Notes (1)
- probe and plasmid building (58)
- Project Meeting (3)
- Protocols (13)
- Research Planning (74)
- Seminars (21)
- Shadow Enhancers (59)
- snail patterning (40)
- Software Development (5)
- Summaries (1,412)
- Teaching (9)
- Transcription Modeling (40)
- Uncategorized (10)
- Web development (19)
Links
Tags
analysis cell culture cell labeling chromatin cloning coding communication confocal data analysis embryo collection embryo labeling figures fly work genomics hb image analysis image processing images in situs Library2 literature making antibodies matlab-storm meetings modeling MP12 mRNA counting Ph planning presentation probe making project 2 project2 result results sectioning section staining shadow enhancers sna snail staining STORM STORM analysis troubleshooting writing-
GitHub Projects
Monday 05/18/15
9:45 am
Chromatin Paper
- writing first draft of letter to editor
- working on probe table
- still need to make primer table.
MERFISH
- working on revised analysis pipeline
Personal stuff
- looking at apartments
Posted in Summaries
Comments Off on Monday 05/18/15
Sunday 05/17/15
11:00 am – 1:00 am
Chromatin manuscript
- computed p-values for overlap figures. Added values to fig caption
- maybe should go into main text when I get it back from Jeff
- p-values for overlap fraction for reference
- Repressed-Active interact less than Silent-Active, p < 1e-14.
- repressed intradomain interact more than active, p < 1e-26.
- repressed intradomain interactions more than silent, p < 1e-22.
- silent intradomain interact more than active, p < 1e-2.
- silent active interact less than silent-silent, p < 1e-10.
- p-values for entanglement score for reference
- Repressed-Active interact less than Silent-Active, p < 1e-4 (~1e-5)
- repressed intradomain interact more than active, p < 1e-12.
- repressed intradomain interactions more than silent, p < 1e-9.
- silent intradomain interact more than active, p = .1 (Not signficant)
- silent active interact less than silent-silent, p < 1e-4
- updated linker colors in Extended Data Fig 6
- computing p-values for for extended data fig 8
Posted in Summaries
Comments Off on Sunday 05/17/15
Friday 05/15/15
9:30 am – 11:55 pm
New Data analysis tools
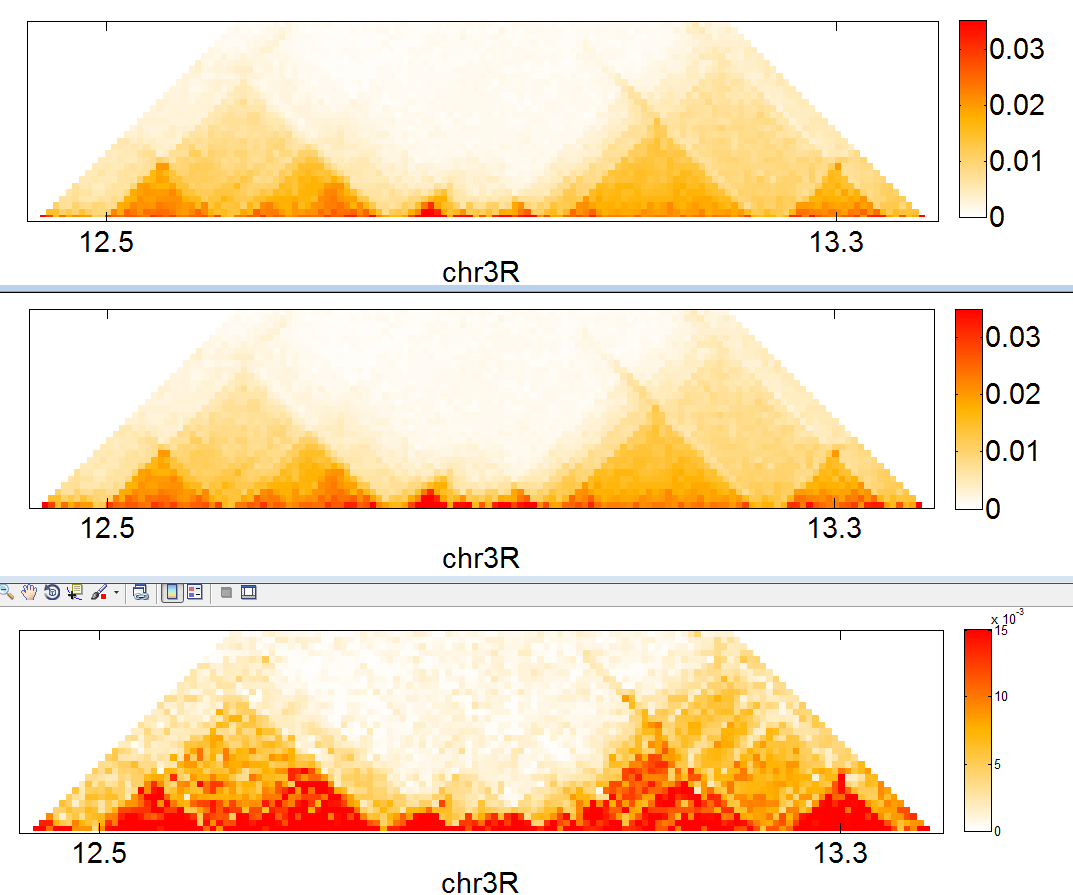
- generalized Hi-C plotter with Bogdan
- now does triangles too
- uploaded new HiC data. Looks much smoother, replicates with different enzymes look very similar.
Chromatin paper
- working on manuscript revisions
- discussed plans and progress with LM and team (6pm-8:30pm)
To do still
- comment on cell cycle and pairing in intro section (may need additional supplemental discussion)
- add p-values to all comparisons in Fig 2, 3, and Extended Data Fig 8 (and 6 and 7), with N cells per comparison.
- XZ recommends combining samples of same type for the fig 2/3/6/7 comparisons.
- check entanglement scores (rerun?)
- fix connectors
- combined fasta file
Posted in Summaries
Comments Off on Friday 05/15/15
Thursday 05/14/15
9:00 am – 8:30 pm
morning 9 – 10:45am
- chat with HC
- email Geoff, congrats and follow up
- email DB about potential collaboration
- create 1 page pdf of recent publication info for K99 update
Chromatin paper methods revision
- working on description of domain selection, 10:45 am – ?
Posted in Summaries
Comments Off on Thursday 05/14/15
Wednesday 05/13/15
9:00 am – 10:15 pm
Geoff’s Defense (9:30 am – 11:00 am)
- see post
- short follow-up discussion with BB and GN
To Do
- Travel reimbursements
- write to XZ about LM travel schedule (done)
Chromatin Paper
- Built new composite data structure (allData.matb) to avoid mixed data sets and crazy indexing.
- some manual field alignment: print elements to Excel, ID missing fields, annotate back in by hand.
- built new domainData2 structure which has the library number and positions (just positions was getting very confusing)
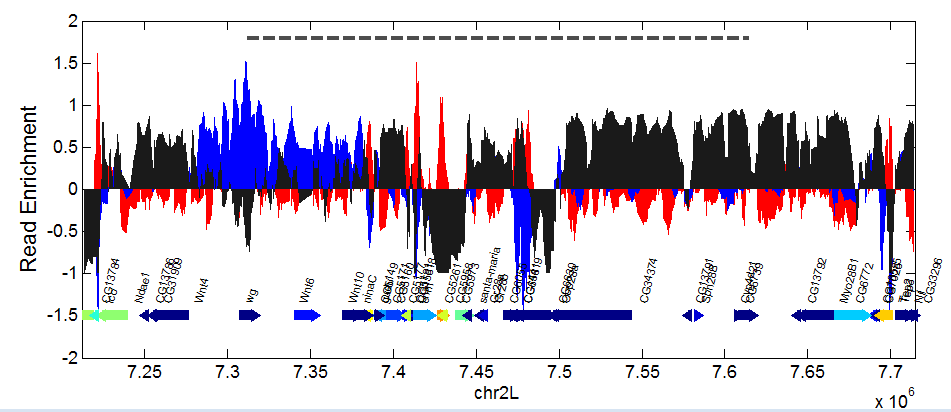
- Completed assembly of Supplemental Figure on Intact Domain Histograms
Extended data Figure revisions (3pm – 5pm)
- renumbering extended data figures in Main text, figures, and figure captions.
- Writing new extended data figure captions.
New Extended Data Figure 8:
- wrote generic bootstrapping function to compute confidence intervals.
- set confidence intervals to 78% for errorbars, So probability two data points with confidence intervals that just overlap are not different ~.22^2 = 0.05.
- alternatively could do the traditional SEM and the error bars would be smaller still.
Still to do
- Write cover letter
- Write description of domain selection
- color connecting bars in Fig E6
- print multiple versions of green/magenta/white contrast for Fig 2 and 3
- combine probe fasta tables and update probe names with new conventions
MERFISH-lunch
- Jeff will send script on new probe design for human genome
- Try building a matlab class to handle library design?
Posted in Summaries
Comments Off on Wednesday 05/13/15
Geoff Fudenberg Thesis Defense
Thesis Defense in Biophysics 05/13/15
Intro
- many different text book picutres of mitotic chromosome
- middle scale: rosettes? spirals of spirals? Accordion coils?
- many functional characterizations of the 1D genome sequence
- Hi-C method explained.
- had to develop appropriate normalization pipeline
- now have maps for many species
Observations
- human contact maps (2009) — 100 fold difference in lengths, see scaling exponent ~1 across this range.
- conclusion chromosomes are polymers
- two compartments.
- year 1 of Geoff’s PhD
- TADs
cycle dependent differences
- metaphase more gentle slope (longer range contacts)
- more homogeneous
- (3 cell types examined)
- locus specific organization lost in metaphase
- cell type specific contacts (there certainly are some) are largely lost
- not evident in the contact range map, but didn’t expect that.
- distinguish between hierarchical models (e.g. coils of coils). and loop array models for metazoan chromosmes
Polymer models
- basic polymer linked monomers
- add model features to test.
- arrays of loops
- confinement
- locus specific interactions
- loops of loops model (hierarchical distinguishable fiber — contact probability vs. distance decays rapidly (more so than observed in metaphase)
- can find scaffold maps that produce this, IF positions of loops are variable from cell-to-cell.
- loops not formed by dimerization. Formed by linking consecutive marks
- random looping model not consistent with HiC data
- Cohesin complexes have the ability to form loops and extrude the DNA through the cohesin loop.
- see citations.
- Q: Comment more what is known about the mechanism of loop extrusion.
- add multiAT hook protein to xenopus chromatin and this condenses into a long shape.
Interphase organization
- compartments vs. domains
- domains are linked to regulation (guide regulatory elements to genes)
- domains inhibit contacts across domains
- Hi resolution Hi-C loops/peaks-at-corners diversity in domain structure + complex domains / loops within loops
- could loop extrusion and boundaries to loop extrusion give rise to interphase domain organization?
- Model
- factors can bind, exchange with solvent, pump DNA
- location bound is stochastic (?) — would it be averaged out across cells. ??
- model results
- can give rise to complex domains,
- can you walk us through the intuition / loop combinations that give rise to one of these complex domains?
- does multiloop / loops-within loops require
- boundary deletion and spreading (cite Nora 2012) – re-examine this.
- loop extrusion more like to care about direction of sites (in-pointing CTCF cites).
Questions
- boundaries block extrusion at certain points along chromsome.
- allow nested loops. (loops stacked completely together).
- Kleckner — what if loops tend to form by collision, in a way limited by persistence length of fiber.
- extrusion is an interesting explanation for how a 1D boundary becomes a 3D boundary.
- well, really all this gives you in nearest boundary element finding.
- not sure this works. Internal interaction within a domain?
- is extrusion
Posted in Seminars
Comments Off on Geoff Fudenberg Thesis Defense
Tuesday 05/12/15
Chromatin Paper
- updated PlotGenes to fix issues with arrowheads. Much nicer now.
- applied in Extended Data Fig 3. Others still need updating.
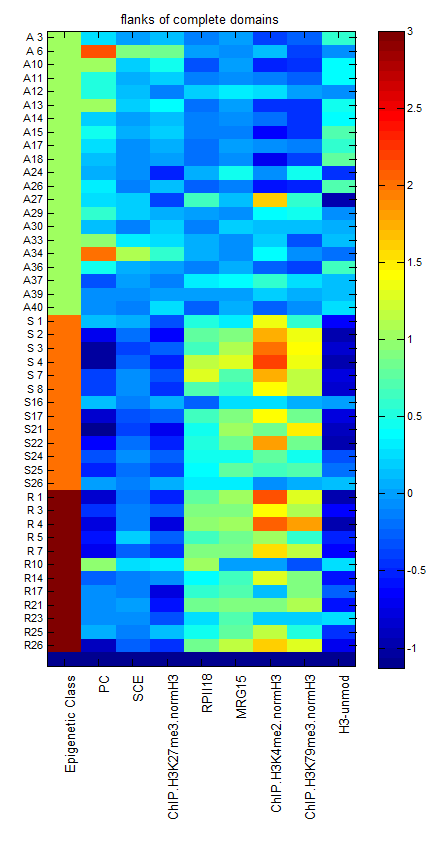
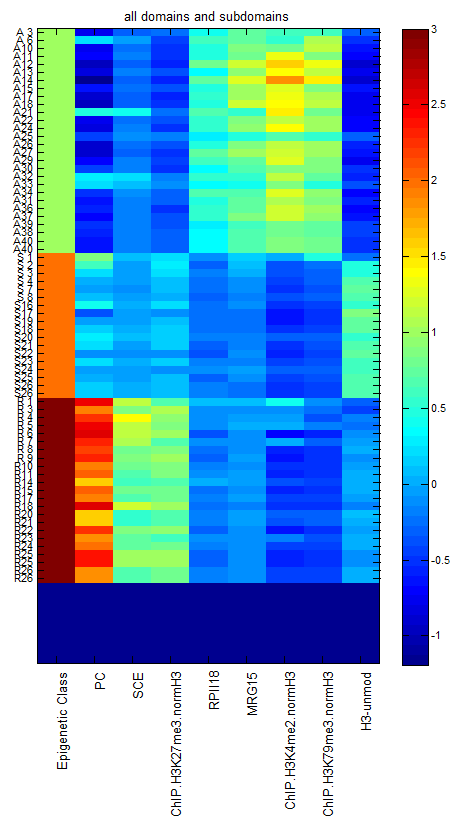
- Working on Extended Data Fig 6: illustrating differences between active domains of similar size.
Paper To Do list
Text
- For the abstract and mostly intro: don’t over-emphasize epigenetic structure. We study structure at the relevant length scales for gene regulation / genome functions. Epingenetic domains are one of the indicators that this length scale is important, and it turns out a strong predictor of structure.
- With respect to Blue internal scaling, can’t say most models — invites question of other models. Move to discussion of models, not in data. Discuss the equilibrium globule model. Say no model fits all the data for Blue.
- Need to discuss off-trend points in model section
- be sure word “loop” does not appear in text
- need to write Cover Letter.
Main Figures
- add scalebars to Fig 1a
- need multiple examples of different contrast /saturation options for Fig 2a and b
- contrast / saturation for Repressed domains is a bit strong.
- images are a little blury — plot with different Gaussian widths
Extended Data Figure
- New Fig
- histograms of volume and radius of gyration distributions for all domains.
- similar histograms for FG model data?
- ED2 ‘internal scaling of repressed regions’
- reorder panels. BX-C vol, ANTC chip, ANTC rg, ANTC vol
- All int data plots: solid lines for int-data, dashed for old data
- Tracing fig
- no backbone
- 15 spots
- green-green junctions linked by green lines. magenta by magenta lines. Black lines between interjunctions
- also in data follow junction code.
- New example stats
- select regions similar length
- show existing stats
- also show cell-cell variation
Posted in Summaries
Comments Off on Tuesday 05/12/15
Monday 05/11/15
9:00 am – 9:50 pm
Chromatin Manuscript
- revising figures and color balance
- helping XZ images and slides
Other stuff
- discussed projects with BB
- discussed L12 progress with JM and HC
- helping GN troubleshoot dax readers
Posted in Summaries
Comments Off on Monday 05/11/15
Protected: Biology of Genomes 2015: Sat Morning (non-open talks)
Posted in Conference Notes
Comments Off on Protected: Biology of Genomes 2015: Sat Morning (non-open talks)