November 2025 M T W T F S S « Aug 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 Categories
- AP patterning (13)
- Blog (1)
- Chromatin (88)
- Conference Notes (72)
- Fly Work (54)
- General STORM (25)
- Genomics (134)
- Journal Club (22)
- Lab Meeting (66)
- Microscopy (79)
- Notes (1)
- probe and plasmid building (58)
- Project Meeting (3)
- Protocols (13)
- Research Planning (74)
- Seminars (21)
- Shadow Enhancers (59)
- snail patterning (40)
- Software Development (5)
- Summaries (1,412)
- Teaching (9)
- Transcription Modeling (40)
- Uncategorized (10)
- Web development (19)
Links
Tags
analysis cell culture cell labeling chromatin cloning coding communication confocal data analysis embryo collection embryo labeling figures fly work genomics hb image analysis image processing images in situs Library2 literature making antibodies matlab-storm meetings modeling MP12 mRNA counting Ph planning presentation probe making project 2 project2 result results sectioning section staining shadow enhancers sna snail staining STORM STORM analysis troubleshooting writing-
GitHub Projects
Protected: chromatin results: 09/05/14
Posted in Chromatin
Comments Off on Protected: chromatin results: 09/05/14
Protected: project 2: update 09/04/14
Posted in Genomics
Comments Off on Protected: project 2: update 09/04/14
Thursday 09/04/14
10:00 am – 11:30 pm
Project 2
- see notes
- (mostly a tedious debugging session).
Chromatin Project, 3D interaction analysis
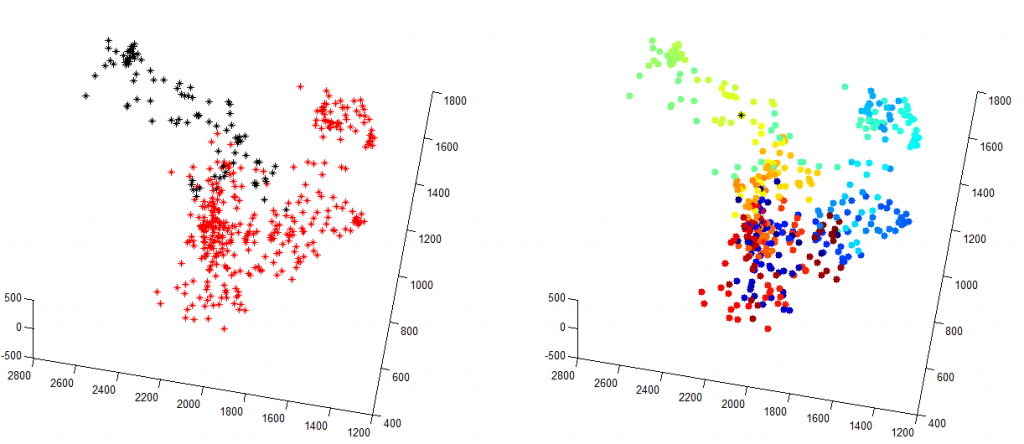
Shown left is the the two domains color coded.
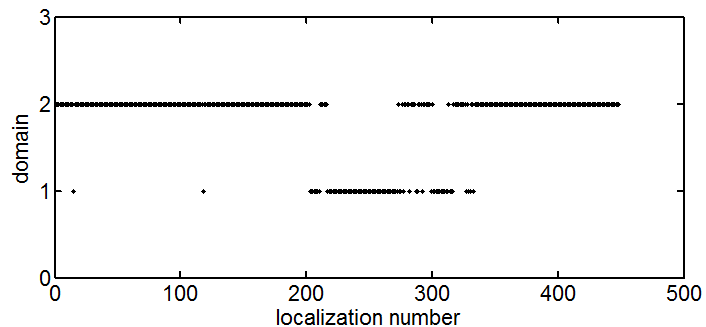
Shown right, I’ve concatenated all the points from both molecule lists, and then plotted the shortest path through all points (travelling-salesman problem solution), with color representing order visited. As you can see, it does a pretty good job of separating these two domains.
My TS solution is annoying slow, nor do I have a good metric yet for ‘converged’. In the images above I’ve downsampled the original data 10 fold so that it completes in a reasonable time. Given that it’s NP-hard I should also probably not be trying to do it in matlab, there must be a fast implementation of this in C…
We then want to turn this into a simple scalar quantification of entanglement.
I think this can be done by looking at how often the TS-route switches between colors. If they are completely separate domains, it should start in one color and finish in the other. As the domains become more entangled, there will be more places where it switches back and forth between them.
Still, if we knew the location of one of the distal ends of the chromatin that we could use as a start point for the route. Then if we also knew we had only a single polymer fiber (rather than a homolog bundle) and if our localization density was high enough, we we could have a decent guess as reconstructing some of the polymer topology. e.g. something like:
Should try again with Fiji 3D multicolor rendering.
Posted in Summaries
Comments Off on Thursday 09/04/14
Wednesday 09/03/14
10:00 am – 10:00 pm
STORM
- troubleshooting storm2 lags with hazen
- might be due to old settings and software on the Hamamatsu “demo” computer creating issues.
- Hazen also troubleshoot issues with the settings file.
- set up STORM of L4E7. lag gets worse with longer imaging sessions.
data analysis with Bogdan
- D8to12 agrees very nicely.
- Bogdan’s more recently analyzed double regions fall quite close to the expected positions for linear scaling.
- singles should get reprocessed with smaller box size and/or more aggressive cut-off against background.
PH paper
- send back thoughts on reviewers to recommend
- review XZ changes to supp mat, send comments to NF
- caught a few tiny typos in the supp mat other sections, need to send these to NF as well
- to do: review cover letter, send comments to NF
Posted in Summaries
Comments Off on Wednesday 09/03/14
Tuesday 09/02/14
10:00 am – 11:30 pm
Chromatin Data Analysis
- troubleshooting failed drift correction using auto-correlation
- reprocessed all recent 2 color data
- working on new method of quantifying overlap — just average distance to all self points vs average distance to all flanking domain points. If this is near 1, the domains are highly entangled.
- should probably normalize this by radius of gyration.
Data Observations
- started exporting 3-color maps of blue regions for figures
- all my current blue regions are flanked on both sides by yellow chromatin. No Blue-black.
Important control
- 2D projection estimates of radius of gyration may be systematic underestimates
Paperwork
- uploaded final version of BWF application
STORM
- attempting imaging of chromatin L4E7
- some cells still have 2 spots, occasionally also satellites. Most look more 1 spot as expected.
- should still quantify, not sure its 80% paired, probably leaning that way though.
- major lag issues with hal recording
- dave bleaching issue does not appear to be reproducible
- had system in true TIRF for a while, 647 spots inside nuclei completely invisible (all cells/nuclei 647 stain data completely invisible, had me confused that I lost cells for a while).
- cells a bit sparse, I may have missed the polyLysine coating on these guys.
Posted in Summaries
Comments Off on Tuesday 09/02/14
Monday 09/01/14
10:00 am – 3:00 pm, 10:45 pm — 12:45 am
Chromatin project
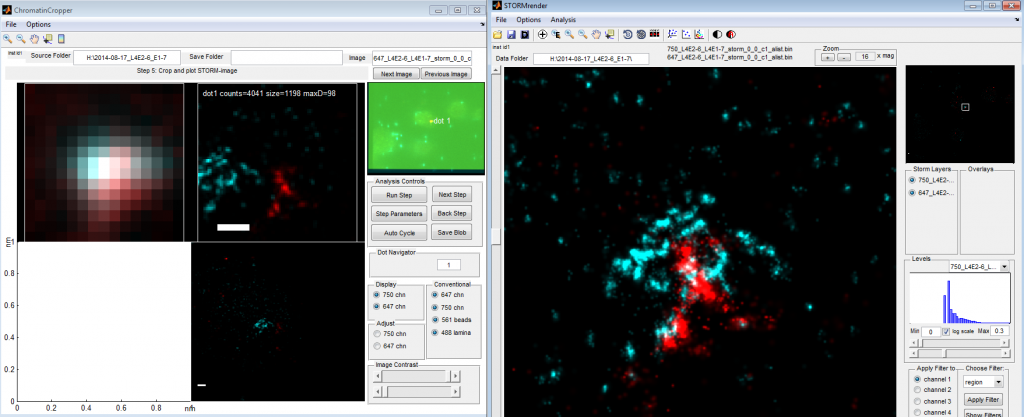
- working on multi-channel global drift correction
- single channel looks great. STORMrender works, but ChromatinCropper has a shift.
- Fixed bug (flipped sign) in ChromatinCroppers Multi-channel drift correction. Now good.
- start redoing recent ChromatinCropper multichannel spot analysis.
New Stains
- (G10 regions, repeat stain)
- L4 E7
- L4 E8
- L4 E6-10
Tasks
- BW application form sent to XZ.
- got signature, need to upload!
Posted in Summaries
Comments Off on Monday 09/01/14
Protected: project 2, planning
Posted in Genomics, Research Planning
Comments Off on Protected: project 2, planning
Sunday 08/31/14
10:00 am – 11:30 pm
Project 2
- see post
Chromatin
- need to fix multichannel drift correction in new version of ChromatinCropper
Posted in Summaries
Comments Off on Sunday 08/31/14
Protected: project 2 update:08/31/14
Posted in Genomics
Comments Off on Protected: project 2 update:08/31/14
Protected: chromatin draft figures
Posted in Chromatin
Comments Off on Protected: chromatin draft figures