rRNA continued
- Goals: do we have all the abundant rRNA transcripts annotated and assembled into our fasta file for screening?
- Annotate FPKMs into list of rRNA and tRNA
more stupid issues not getting the right genes
There are yeast genes in my list of all human rRNAs.
lcl|IsoformID:_ENST00000544484.1__GeneID:_ENSG00000111642.10__FPKM:_2000000__name:_CHD4__locus:_chr12:6679248-6716534
AADB02025922.6474.8212 Eukaryota;Opisthokonta;Holozoa;Metazoa;Animalia;Craniata;Mammalia;
lcl|IsoformID:_ENST00000544484.1__GeneID:_ENSG00000111642.10__FPKM:_2000000__name:_CHD4__locus:_chr12:6679248-6716534
AC136932.58798.60652 Eukaryota;Opisthokonta;Holozoa;Metazoa;Animalia;Craniata;Mammalia;
lcl|IsoformID:_ENST00000544484.1__GeneID:_ENSG00000111642.10__FPKM:_2000000__name:_CHD4__locus:_chr12:6679248-6716534
AC138525.467347.469136 Eukaryota;Opisthokonta;Nucletmycea;Fungi;Dikarya;Ascomycota;Saccharo mycotina;Saccharomycetes;Saccharomycetidae;Saccharomycetales;Sacchar omycetaceae;Saccharomyces;
lcl|IsoformID:_ENST00000544484.1__GeneID:_ENSG00000111642.10__FPKM:_2000000__name:_CHD4__locus:_chr12:6679248-6716534
ADDF02155034.50.1904 Eukaryota;Opisthokonta;Holozoa;Metazoa;Animalia;Craniata;Mammalia;
lcl|IsoformID:_ENST00000544484.1__GeneID:_ENSG00000111642.10__FPKM:_2000000__name:_CHD4__locus:_chr12:6679248-6716534
ABBA01017804.504.2358 Eukaryota;Opisthokonta;Holozoa;Metazoa;Animalia;Craniata;Mammalia;
lcl|IsoformID:_ENST00000544484.1__GeneID:_ENSG00000111642.10__FPKM:_2000000__name:_CHD4__locus:_chr12:6679248-6716534
ABSL01023168.504.2358 Eukaryota;Opisthokonta;Holozoa;Metazoa;Animalia;Craniata;Mammalia;Ho mo sapiens (human)
lcl|IsoformID:_ENST00000544484.1__GeneID:_ENSG00000111642.10__FPKM:_2000000__name:_CHD4__locus:_chr12:6679248-6716534
AC139250.551258.553047 Eukaryota;Opisthokonta;Nucletmycea;Fungi;Dikarya;Ascomycota;Saccharo mycotina;Saccharomycetes;Saccharomycetidae;Saccharomycetales;Sacchar omycetaceae;Saccharomyces;
lcl|IsoformID:_ENST00000544484.1__GeneID:_ENSG00000111642.10__FPKM:_2000000__name:_CHD4__locus:_chr12:6679248-6716534
AMYH02031946.191090.192944 Eukaryota;Opisthokonta;Holozoa;Metazoa;Animalia;Craniata;Mammalia;
lcl|IsoformID:_ENST00000544484.1__GeneID:_ENSG00000111642.10__FPKM:_2000000__name:_CHD4__locus:_chr12:6679248-6716534
AC138524.219038.220826 Eukaryota;Opisthokonta;Nucletmycea;Fungi;Dikarya;Ascomycota;Saccharo mycotina;Saccharomycetes;Saccharomycetidae;Saccharomycetales;Sacchar omycetaceae;Saccharomyces;
lcl|IsoformID:_ENST00000544484.1__GeneID:_ENSG00000111642.10__FPKM:_2000000__name:_CHD4__locus:_chr12:6679248-6716534
FP885865.117573.119427 Eukaryota;Opisthokonta;Holozoa;Metazoa;Animalia;Craniata;Mammalia;
lcl|IsoformID:_ENST00000426789.1__GeneID:_ENSG00000072274.8__FPKM:_2000000__name:_TFRC__locus:_chr3:195754053-195782085
Homo_sapiens_chr2.trna6-GluTTC (75124046-75124114) Glu (TTC) 69 bp Sc: 23.54
lcl|IsoformID:_ENST00000368445.5__GeneID:_ENSG00000160691.14__FPKM:_2000000__name:_SHC1__locus:_chr1:154934773-154943217
Homo_sapiens_chr6.trna155-LeuTAA (27198416-27198334) Leu (TAA) 83 bp Sc: 74.34
lcl|IsoformID:_ENSG00000156471.8__GeneID:_ENSG00000156471.8__FPKM:_2000000__name:_PTDSS1__locus:_chr8:97273942-97349223
Homo_sapiens_chr2.trna25-GluCTC (71273560-71273488) Glu (CTC) 73 bp Sc: 22.62
I am not convinced I have rRNAs in the total RNA dat from the IMR90s from ENCODE. My top expressed RNAs from that list have more ribosomal proteins than ribosomal RNAs.
All FPKMs over 1 million
'IsoformID:_ENST00000426789.1__GeneID:_ENSG00000072274.8__FPKM:_34000000__name:_TFRC__locus:_chr3:195754053-195782085'
'IsoformID:_ENST00000374875.1__GeneID:_ENSG00000099250.12__FPKM:_30000000__name:_NRP1__locus:_chr10:33466419-33623564'
'IsoformID:_ENST00000489283.1__GeneID:_ENSG00000102024.12__FPKM:_22000000__name:_PLS3__locus:_chrX:114795500-114864228'
'IsoformID:_ENST00000544484.1__GeneID:_ENSG00000111642.10__FPKM:_32000000__name:_CHD4__locus:_chr12:6679248-6716534'
'IsoformID:_ENSG00000122729.13__GeneID:_ENSG00000122729.13__FPKM:_6000000__name:_ACO1__locus:_chr9:32384617-32450834'
'IsoformID:_ENST00000463004.1__GeneID:_ENSG00000131051.16__FPKM:_88000000__name:_RBM39__locus:_chr20:34291530-34330158'
'IsoformID:_ENSG00000156471.8__GeneID:_ENSG00000156471.8__FPKM:_14000000__name:_PTDSS1__locus:_chr8:97273942-97349223'
'IsoformID:_ENSG00000157654.13__GeneID:_ENSG00000157654.13__FPKM:_8000000__name:_PALM2-AKAP2__locus:_chr9:112542588-112934792'
'IsoformID:_ENST00000368445.5__GeneID:_ENSG00000160691.14__FPKM:_26000000__name:_SHC1__locus:_chr1:154934773-154943217'
'IsoformID:_ENST00000368457.2__GeneID:_ENSG00000163348.3__FPKM:_8000000__name:_PYGO2__locus:_chr1:154929501-154934224'
'IsoformID:_ENSG00000175106.12__GeneID:_ENSG00000175106.12__FPKM:_20000000__name:_TVP23C__locus:_chr17:15341204-15466909'
'IsoformID:_ENST00000438150.2__GeneID:_ENSG00000198538.6__FPKM:_20000000__name:_ZNF28__locus:_chr19:53300661-53309274'
'IsoformID:_ENST00000362862.1__GeneID:_ENSG00000199732.1__FPKM:_2000000__name:_Y_RNA__locus:_chr8:97321448-97321550'
'IsoformID:_ENST00000595646.1__GeneID:_ENSG00000204604.5__FPKM:_16000000__name:_ZNF468__locus:_chr19:53341260-53360872'
'IsoformID:_ENSG00000223878.1__GeneID:_ENSG00000223878.1__FPKM:_2000000__name:_AC005517.3__locus:_chr17:15410179-15410668'
'IsoformID:_ENST00000418441.1__GeneID:_ENSG00000228053.1__FPKM:_2000000__name:_RP11-151F5.2__locus:_chr9:112787160-112787638'
'IsoformID:_ENSG00000228413.1__GeneID:_ENSG00000228413.1__FPKM:_4000000__name:_AC024937.2__locus:_chr3:195771565-195772125'
'IsoformID:_ENSG00000232939.1__GeneID:_ENSG00000232939.1__FPKM:_4000000__name:_RP11-406O23.2__locus:_chr9:112522639-112534323'
'IsoformID:_ENST00000424673.1__GeneID:_ENSG00000234324.1__FPKM:_2000000__name:_RPL9P2__locus:_chr17:15346952-15347505'
'IsoformID:_ENSG00000238258.1__GeneID:_ENSG00000238258.1__FPKM:_4000000__name:_RP11-342D11.2__locus:_chr10:33500204-33502732'
'IsoformID:_ENST00000459110.1__GeneID:_ENSG00000238549.1__FPKM:_2000000__name:_snoU13__locus:_chr20:34304392-34304492'
'IsoformID:_ENST00000312177.6__GeneID:_ENSG00000239704.6__FPKM:_10000000__name:_CDRT4__locus:_chr17:15339331-15370925'
'IsoformID:_ENST00000434623.2__GeneID:_ENSG00000241978.5__FPKM:_16000000__name:_AKAP2__locus:_chr9:112810877-112932189'
'IsoformID:_ENST00000374531.2__GeneID:_ENSG00000243444.3__FPKM:_14000000__name:_PALM2__locus:_chr9:112403067-112708983'
'IsoformID:_ENSG00000247853.2__GeneID:_ENSG00000247853.2__FPKM:_4000000__name:_RP5-940J5.6__locus:_chr12:6687787-6693905'
'IsoformID:_ENSG00000250182.2__GeneID:_ENSG00000250182.2__FPKM:_2000000__name:_EEF1A1P13__locus:_chr5:14652046-14653438'
'IsoformID:_ENST00000516365.1__GeneID:_ENSG00000252174.1__FPKM:_2000000__name:_RNU7-18P__locus:_chr3:195799689-195799751'
'IsoformID:_ENSG00000252441.1__GeneID:_ENSG00000252441.1__FPKM:_2000000__name:_SNORA64__locus:_chrX:114779968-114780049'
'IsoformID:_ENST00000536597.2__GeneID:_ENSG00000255875.2__FPKM:_2000000__name:_CTD-2102P23.1__locus:_chr19:53313076-53313961'
'IsoformID:_ENST00000580625.1__GeneID:_ENSG00000258486.2__FPKM:_1849422__name:_RN7SL1__locus:_chr14:50053297-50053594'
'IsoformID:_ENSG00000259024.2__GeneID:_ENSG00000259024.2__FPKM:_10000000__name:_TVP23C-CDRT4__locus:_chr17:15339337-15466875'
'IsoformID:_ENST00000490232.2__GeneID:_ENSG00000265150.1__FPKM:_1097220__name:_RN7SL2__locus:_chr14:50329270-50329567'
'IsoformID:_ENST00000605085.1__GeneID:_ENSG00000271380.1__FPKM:_2000000__name:_RP11-307C12.12__locus:_chr1:154934300-154935099'
'IsoformID:_ENSG00000271826.1__GeneID:_ENSG00000271826.1__FPKM:_6000000__name:_RP1-93I3.1__locus:_chrX:114752489-114797058'
contains Y_RNAs and snoRNAs so it’s clearly not just polyAs. Lots of RNA binding proteins too.
More issues !
why are there more total isoforms in the polyA cufflinks than in the totRNA output?
length(polyaRNA) = 230312
length(totRNA) = 229007
however by comparing gene names in the isoform list I get all the same hits. so both lists are complete in terms of genes.
looking at just shared transcripts. Enriched 1000 fold and FPKMs over 1000
‘HIST2H4B’
‘HIST1H4C’
‘HIST1H4L’
‘SNORD9’
‘Y_RNA’
‘RNU5A-1’
‘SNORA52’
‘SNORA73B’
‘RNU5B-1’
‘RNU5D-1’
‘SNORA63’
‘Y_RNA’
‘Y_RNA’
‘Y_RNA’
‘SNORD8’
‘RNU4-1’
‘SNORA38’
‘Y_RNA’
‘RNU5A-8P’
‘RN7SKP71’
‘RNU4-91P’
‘RNY3P1’
‘Y_RNA’
‘RNY3’
‘SNORA57’
‘SNORA5A’
‘RNU1-2’
‘SNORA7B’
‘Y_RNA’
‘SNORA70’
‘SNORA64’
‘RNU6-15P’
‘Y_RNA’
‘SNORA8’
‘RNU1-4’
‘SNORD15B’
‘SNORA7A’
‘SNORA48’
‘RNU6ATAC2P’
‘SNORA34’
‘RNU2-2P’
‘RNU2-59P’
‘TMSB4XP6’
‘EEF1A1P6’
‘RPL39P3’
‘SNORD10’
‘RN7SL521P’
‘RN7SL364P’
‘SCARNA22’
‘SCARNA1’
‘RNU4ATAC5P’
‘SNORA31’
‘RPPH1’
‘SNORD3A’
‘RNU4ATAC’
‘RMRP’
‘SCARNA2’
‘RNU11’
‘SNORA51’
‘SNORA28’
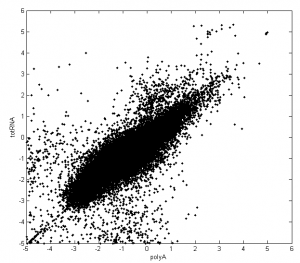
Morre on missing genes:
There are highly expressed genes in the polyA data that aren’t in the totRNA data:
highMissed =
'MT-RNR2' '2068.38'
'MT-CO1' '7390.12'
'MT-CO2' '5739.78'
'MT-ATP8' '10564.1'
'MT-ATP6' '1815.62'
'MT-CO3' '1863.87'
'MT-ND4L' '10204.1'
'MT-ND4' '2253.71'
'MT-ND5' '1125.1'
It appears these are the mitochondrial genes? Nothing in ENCODE leads me to expect that the mitochondrial genes were excluded from the totRNA but included in the polyA… This could have been annotated better.
Analysis of outstanding libraries
- L4E2 clusters look a bit better
L4E3
Issues with top found genes:
These are not trivially the rRNA targets:
‘L4 E2_LowEx_rep ab20_849 B08 VCP ENST00000493886.1 681 B15 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B03 PSMB4 ENST00000290541.6 648 B16 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B04 PHB2 ENST00000399433.2 843 B07 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B02 CLIC1 ENST00000375784.3 971 B05 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B03 PSMB7 ENST00000259457.3 861 B08 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B03 HSP90AA1 ENST00000216281.8 750 B05 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B02 ID3 ENST00000374561.5 329 B16 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B07 TMEM147 ENST00000222284.5 340 B09 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B01 TAGLN2 ENST00000368097.4 335 B07 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B12 SNRPB ENST00000381342.2 623 B14 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B10 NCAPG2 ENST00000275830.10 317 B13 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B02 CERS5 ENST00000317551.6 232 B10 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B03 CERS5 ENST00000317551.6 201 B10 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B11 MAD2L2 ENST00000376692.4 922 B13 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B02 LRP11 ENST00000546019.1 413 B03 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B10 PABPC1L ENST00000217074.4 785 B13 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B05 TDG ENST00000392872.3 870 B13 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B08 PIM3 ENST00000360612.4 178 B12 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B09 PIM3 ENST00000360612.4 258 B12 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B01 BAD ENST00000309032.3 318 B06 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B13 ARRDC1 ENST00000371421.4 971 B16 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B04 SCYL1 ENST00000270176.5 602 B14 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B01 H19 ENST00000414790.1 609 B04 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B02 PBX1 ENST00000367897.1 225 B12 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B03 PBX1 ENST00000367897.1 166 B04 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B09 RELB ENST00000505236.1 773 B10 ab20_1492’
‘L4 E2_LowEx_rep ab20_849 B03 E.Coli_8 ENS_E.Coli_8 971 B08 ab20_1492’
Probes that align to highly expressed non-PolyA sequences:
ans =
'L4 E2_LowEx_rep ab20_849 B05 VCP ENST00000493886.1 727 B08 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B01 CLIC1 ENST00000375784.3 755 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B13 PDXK ENST00000343528.6 227 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 S100A16 ENST00000368706.4 221 B08 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B12 SNRPB ENST00000381342.2 717 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B06 MAD2L2 ENST00000376692.4 76 B13 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 PDE6D ENST00000287600.4 179 B13 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B05 TDG ENST00000392872.3 283 B07 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B01 PIM3 ENST00000360612.4 510 B09 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B09 MAP3K11 ENST00000309100.3 117 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B14 MAP3K11 ENST00000309100.3 320 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B05 E.Coli_12 ENS_E.Coli_12 921 B15 ab20_1492'
ans =
'lcl|IsoformID:_ENST00000368457.2__GeneID:_ENSG00000163348.3__FPKM:_8000000__name:_PYGO2__locus:_chr1:154929501-154934224'
'lcl|IsoformID:_ENST00000424673.1__GeneID:_ENSG00000234324.1__FPKM:_2000000__name:_RPL9P2__locus:_chr17:15346952-15347505'
'lcl|IsoformID:_ENST00000368457.2__GeneID:_ENSG00000163348.3__FPKM:_8000000__name:_PYGO2__locus:_chr1:154929501-154934224'
'lcl|IsoformID:_ENST00000424573.1__GeneID:_ENSG00000235174.1__FPKM:_1153.31__name:_RPL39P3__locus:_chr6:74082830-74082986'
'lcl|IsoformID:_ENSG00000222626.1__GeneID:_ENSG00000222626.1__FPKM:_1421.94__name:_RNU2-48P__locus:_chr5:157403773-157403964'
'lcl|IsoformID:_ENST00000605085.1__GeneID:_ENSG00000271380.1__FPKM:_2000000__name:_RP11-307C12.12__locus:_chr1:154934300-154935099'
'lcl|IsoformID:_ENST00000386847.1__GeneID:_ENSG00000209582.1__FPKM:_1955.56__name:_SNORA48__locus:_chr17:7478030-7478165'
'lcl|IsoformID:_ENST00000368457.2__GeneID:_ENSG00000163348.3__FPKM:_8000000__name:_PYGO2__locus:_chr1:154929501-154934224'
'lcl|IsoformID:_ENSG00000238258.1__GeneID:_ENSG00000238258.1__FPKM:_4000000__name:_RP11-342D11.2__locus:_chr10:33500204-33502732'
'lcl|IsoformID:_ENST00000605085.1__GeneID:_ENSG00000271380.1__FPKM:_2000000__name:_RP11-307C12.12__locus:_chr1:154934300-154935099'
'lcl|IsoformID:_ENST00000368445.5__GeneID:_ENSG00000160691.14__FPKM:_26000000__name:_SHC1__locus:_chr1:154934773-154943217'
'lcl|IsoformID:_ENST00000368457.2__GeneID:_ENSG00000163348.3__FPKM:_8000000__name:_PYGO2__locus:_chr1:154929501-154934224'
None of these are the way over-abundant species in L4E2. Many of the over abundant L4E2s align to genes with lower FPKMs. Maybe these lower FPKMs are under-estimated though by the 200 bp cutoff in the tot RNA.
FPKM over 10
ans =
'L4 E2_LowEx_rep ab20_849 B05 VCP ENST00000493886.1 727 B08 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B08 PSMB4 ENST00000290541.6 148 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B12 PSMB4 ENST00000290541.6 427 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B06 EMC7 ENST00000256545.4 875 B09 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B04 PHB2 ENST00000399433.2 424 B11 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B02 PSMB5 ENST00000361611.6 913 B07 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B04 PSMB5 ENST00000361611.6 18 B06 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B05 TUBA4A ENST00000248437.4 619 B13 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 TK1 ENST00000301634.7 447 B06 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 TK1 ENST00000301634.7 416 B06 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B04 TALDO1 ENST00000319006.3 662 B06 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B06 TALDO1 ENST00000319006.3 326 B13 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B06 UQCRQ ENST00000378670.3 135 B10 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B14 ACTN4 ENST00000588618.1 669 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 CD63 ENST00000550050.1 83 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B13 PDXK ENST00000343528.6 227 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B02 TKT ENST00000423516.1 502 B09 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B07 CCT5 ENST00000503026.1 617 B08 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B07 TIMM50 ENST00000607714.1 965 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B08 TIMM50 ENST00000607714.1 634 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B08 TIMM50 ENST00000607714.1 123 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B05 CLU ENST00000522098.1 506 B07 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 S100A16 ENST00000368706.4 221 B08 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 S100A16 ENST00000368706.4 848 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B12 ERGIC3 ENST00000348547.2 54 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B14 NME4 ENST00000219479.2 209 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B05 CYC1 ENST00000318911.4 402 B06 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 PSMB6 ENST00000270586.3 579 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B01 BLVRB ENST00000263368.4 687 B12 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 SLC25A6 ENST00000381401.5 71 B10 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B04 SLC25A6 ENST00000381401.5 485 B10 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B01 TAGLN2 ENST00000368097.4 206 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B07 TAGLN2 ENST00000368097.4 273 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B07 TAGLN2 ENST00000368097.4 366 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B12 SNRPB ENST00000381342.2 654 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 GNB2L1 ENST00000504325.1 17 B05 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B05 GNB2L1 ENST00000504325.1 823 B10 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B02 LRRFIP1 ENST00000289175.6 389 B11 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B12 ADRBK1 ENST00000308595.5 31 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B02 TUBGCP2 ENST00000417178.2 693 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B05 CTDSP2 ENST00000398073.2 344 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B10 NCAPG2 ENST00000275830.10 813 B13 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B02 CALM3 ENST00000291295.9 940 B09 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B02 CALM3 ENST00000291295.9 603 B09 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B02 CALM3 ENST00000291295.9 65 B10 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B12 UBALD2 ENST00000327490.6 940 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 MRPL49 ENST00000279242.2 156 B05 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B01 RIC8A ENST00000325207.5 312 B02 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B01 RIC8A ENST00000325207.5 854 B12 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B13 PSMD7 ENST00000219313.4 602 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 CERS5 ENST00000317551.6 225 B07 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B02 ANXA11 ENST00000422982.3 321 B09 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B04 ATP6V0A1 ENST00000585828.1 53 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B12 ATP6V0A1 ENST00000585828.1 597 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 PDE6D ENST00000287600.4 179 B13 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B07 PLAUR ENST00000340093.3 27 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B02 LRP11 ENST00000546019.1 618 B09 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B06 C14orf166 ENST00000261700.3 847 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B12 C14orf166 ENST00000261700.3 542 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B11 MMADHC ENST00000303319.5 891 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B10 UBQLN2 ENSG00000188021.7 276 B11 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B05 TDG ENST00000392872.3 283 B07 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B01 PIM3 ENST00000360612.4 510 B09 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B06 SIL1 ENST00000394817.2 378 B08 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B12 ACOT8 ENST00000217455.4 46 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B14 ACOT8 ENST00000217455.4 463 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B01 BAD ENST00000309032.3 869 B06 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B06 BAD ENST00000309032.3 683 B11 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B02 HEXIM1 ENST00000332499.2 725 B07 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B05 CUEDC2 ENST00000369937.4 971 B14 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B10 PTDSS1 ENST00000337004.4 971 B13 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B01 H19 ENST00000414790.1 516 B05 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B04 H19 ENST00000414790.1 749 B05 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B09 MAP3K11 ENST00000309100.3 117 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B09 RELB ENST00000505236.1 618 B13 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B06 E.Coli_1 ENS_E.Coli_1 196 B08 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B07 E.Coli_2 ENS_E.Coli_2 46 B08 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B03 E.Coli_4 ENS_E.Coli_4 15 B11 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B14 E.Coli_6 ENS_E.Coli_6 192 B15 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B12 E.Coli_7 ENS_E.Coli_7 566 B16 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B05 E.Coli_10 ENS_E.Coli_10 847 B06 ab20_1492'
'L4 E2_LowEx_rep ab20_849 B05 E.Coli_12 ENS_E.Coli_12 921 B15 ab20_1492'
ans =
'lcl|IsoformID:_ENST00000368457.2__GeneID:_ENSG00000163348.3__FPKM:_8000000__name:_PYGO2__locus:_chr1:154929501-154934224'
'lcl|IsoformID:_ENST00000462468.2__GeneID:_ENSG00000243488.2__FPKM:_29.0734__name:_RN7SL337P__locus:_chr19:14694906-14695204'
'lcl|IsoformID:_ENSG00000223544.1__GeneID:_ENSG00000223544.1__FPKM:_11.724__name:_AC005838.2__locus:_chr17:15492165-15492398'
'lcl|IsoformID:_ENST00000515982.1__GeneID:_ENSG00000251791.1__FPKM:_23.3477__name:_SCARNA6__locus:_chr2:234197321-234197586'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000432169.1__GeneID:_ENSG00000023228.8__FPKM:_42.2311__name:_NDUFS1__locus:_chr2:206988824-207024149'
'lcl|IsoformID:_ENST00000578793.1__GeneID:_ENSG00000264169.1__FPKM:_293.701__name:_RN7SL665P__locus:_chr9:133275652-133275950'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000294785.5__GeneID:_ENSG00000162736.11__FPKM:_22.8617__name:_NCSTN__locus:_chr1:160313061-160328742'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000541815.1__GeneID:_ENSG00000255749.1__FPKM:_13.3324__name:_GNAI2P1__locus:_chr12:14407845-14408160'
'lcl|IsoformID:_ENST00000583493.1__GeneID:_ENSG00000263424.1__FPKM:_27.0232__name:_CTD-2541J13.2__locus:_chr18:65173825-65181267'
'lcl|IsoformID:_ENST00000494042.2__GeneID:_ENSG00000242101.2__FPKM:_40.61__name:_RN7SL416P__locus:_chr7:100127986-100128282'
'lcl|IsoformID:_ENSG00000199683.1__GeneID:_ENSG00000199683.1__FPKM:_13.599__name:_RN7SKP185__locus:_chr20:36603557-36603883'
'lcl|IsoformID:_ENST00000368457.2__GeneID:_ENSG00000163348.3__FPKM:_8000000__name:_PYGO2__locus:_chr1:154929501-154934224'
'lcl|IsoformID:_ENST00000607615.1__GeneID:_ENSG00000272075.1__FPKM:_114.363__name:_Metazoa_SRP__locus:_chr11:117937951-117938252'
'lcl|IsoformID:_ENST00000241704.7__GeneID:_ENSG00000122218.10__FPKM:_210.6586__name:_COPA__locus:_chr1:160259062-160313190'
'lcl|IsoformID:_ENST00000434133.2__GeneID:_ENSG00000229230.2__FPKM:_223.066__name:_MT1P3__locus:_chr20:33805811-33805931'
'lcl|IsoformID:_ENSG00000240961.2__GeneID:_ENSG00000240961.2__FPKM:_60.4868__name:_RN7SL415P__locus:_chr6:92449326-92449614'
'lcl|IsoformID:_ENST00000416738.1__GeneID:_ENSG00000222024.2__FPKM:_12.6257__name:_AC004945.1__locus:_chr7:78982725-78983708'
'lcl|IsoformID:_ENST00000365536.1__GeneID:_ENSG00000202406.1__FPKM:_17.8056__name:_RN7SKP187__locus:_chr7:111928677-111929007'
'lcl|IsoformID:_ENST00000454464.2__GeneID:_ENSG00000223628.2__FPKM:_10.5899__name:_AC023449.2__locus:_chr15:25826115-25826269'
'lcl|IsoformID:_ENST00000483161.2__GeneID:_ENSG00000240183.2__FPKM:_41.5778__name:_RN7SL297P__locus:_chr2:112687751-112688041'
'lcl|IsoformID:_ENSG00000265053.1__GeneID:_ENSG00000265053.1__FPKM:_47.9133__name:_RN7SL321P__locus:_chr3:48421323-48421620'
'lcl|IsoformID:_ENST00000241704.7__GeneID:_ENSG00000122218.10__FPKM:_210.6586__name:_COPA__locus:_chr1:160259062-160313190'
'lcl|IsoformID:_ENST00000462468.2__GeneID:_ENSG00000243488.2__FPKM:_29.0734__name:_RN7SL337P__locus:_chr19:14694906-14695204'
'lcl|IsoformID:_ENST00000369585.3__GeneID:_ENSG00000126890.13__FPKM:_13.4145__name:_CTAG2__locus:_chrX:153880250-153881842'
'lcl|IsoformID:_ENSG00000263595.1__GeneID:_ENSG00000263595.1__FPKM:_58.1117__name:_RN7SL823P__locus:_chr19:17021302-17021594'
'lcl|IsoformID:_ENST00000306061.6__GeneID:_ENSG00000169715.10__FPKM:_48.4732__name:_MT1E__locus:_chr16:56659386-56661024'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000462960.2__GeneID:_ENSG00000244307.2__FPKM:_58.5404__name:_RN7SL395P__locus:_chr8:146010682-146010971'
'lcl|IsoformID:_ENST00000308018.4__GeneID:_ENSG00000128626.7__FPKM:_13.4189__name:_MRPS12__locus:_chr19:39421187-39423660'
'lcl|IsoformID:_ENSG00000265053.1__GeneID:_ENSG00000265053.1__FPKM:_47.9133__name:_RN7SL321P__locus:_chr3:48421323-48421620'
'lcl|IsoformID:_ENSG00000252636.1__GeneID:_ENSG00000252636.1__FPKM:_144.758__name:_RNU6-826P__locus:_chr17:47113713-47113817'
'lcl|IsoformID:_ENST00000365370.1__GeneID:_ENSG00000202240.1__FPKM:_230.941__name:_RNU6-737P__locus:_chr18:54951832-54951939'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENSG00000265053.1__GeneID:_ENSG00000265053.1__FPKM:_47.9133__name:_RN7SL321P__locus:_chr3:48421323-48421620'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000468562.2__GeneID:_ENSG00000240905.2__FPKM:_54.9867__name:_RN7SL798P__locus:_chr8:56892800-56893096'
'lcl|IsoformID:_ENST00000494719.2__GeneID:_ENSG00000244197.2__FPKM:_19.3915__name:_RN7SL766P__locus:_chr13:22364675-22364982'
'lcl|IsoformID:_ENST00000308018.4__GeneID:_ENSG00000128626.7__FPKM:_13.4189__name:_MRPS12__locus:_chr19:39421187-39423660'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENSG00000265053.1__GeneID:_ENSG00000265053.1__FPKM:_47.9133__name:_RN7SL321P__locus:_chr3:48421323-48421620'
'lcl|IsoformID:_ENST00000241704.7__GeneID:_ENSG00000122218.10__FPKM:_210.6586__name:_COPA__locus:_chr1:160259062-160313190'
'lcl|IsoformID:_ENST00000583493.1__GeneID:_ENSG00000263424.1__FPKM:_27.0232__name:_CTD-2541J13.2__locus:_chr18:65173825-65181267'
'lcl|IsoformID:_ENSG00000218893.1__GeneID:_ENSG00000218893.1__FPKM:_18.1349__name:_RP3-451B15.3__locus:_chr6:12319684-12319824'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000410878.1__GeneID:_ENSG00000222810.1__FPKM:_33.1117__name:_RNU2-68P__locus:_chrX:71596828-71597019'
'lcl|IsoformID:_ENST00000241704.7__GeneID:_ENSG00000122218.10__FPKM:_210.6586__name:_COPA__locus:_chr1:160259062-160313190'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000467650.2__GeneID:_ENSG00000241983.2__FPKM:_225.475__name:_RN7SL566P__locus:_chr19:39859792-39860086'
'lcl|IsoformID:_ENST00000386847.1__GeneID:_ENSG00000209582.1__FPKM:_1955.56__name:_SNORA48__locus:_chr17:7478030-7478165'
'lcl|IsoformID:_ENSG00000240663.2__GeneID:_ENSG00000240663.2__FPKM:_16.3604__name:_RN7SL310P__locus:_chr18:47733369-47733648'
'lcl|IsoformID:_ENSG00000252717.1__GeneID:_ENSG00000252717.1__FPKM:_223.084__name:_RNU6-352P__locus:_chr1:102325406-102325513'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENSG00000265053.1__GeneID:_ENSG00000265053.1__FPKM:_47.9133__name:_RN7SL321P__locus:_chr3:48421323-48421620'
'lcl|IsoformID:_ENSG00000202157.1__GeneID:_ENSG00000202157.1__FPKM:_26.4129__name:_RNU4-11P__locus:_chr5:83099372-83099504'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000368457.2__GeneID:_ENSG00000163348.3__FPKM:_8000000__name:_PYGO2__locus:_chr1:154929501-154934224'
'lcl|IsoformID:_ENSG00000238258.1__GeneID:_ENSG00000238258.1__FPKM:_4000000__name:_RP11-342D11.2__locus:_chr10:33500204-33502732'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENSG00000265053.1__GeneID:_ENSG00000265053.1__FPKM:_47.9133__name:_RN7SL321P__locus:_chr3:48421323-48421620'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000419683.1__GeneID:_ENSG00000226908.1__FPKM:_13.3039__name:_HIST1H2BPS3__locus:_chr13:22057972-22058313'
'lcl|IsoformID:_ENST00000478568.1__GeneID:_ENSG00000153207.10__FPKM:_22.3043__name:_AHCTF1__locus:_chr1:247079615-247095280'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000583493.1__GeneID:_ENSG00000263424.1__FPKM:_27.0232__name:_CTD-2541J13.2__locus:_chr18:65173825-65181267'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000583493.1__GeneID:_ENSG00000263424.1__FPKM:_27.0232__name:_CTD-2541J13.2__locus:_chr18:65173825-65181267'
'lcl|IsoformID:_ENSG00000221852.4__GeneID:_ENSG00000221852.4__FPKM:_17.0181__name:_KRTAP1-5__locus:_chr17:39182277-39183454'
'lcl|IsoformID:_ENST00000605085.1__GeneID:_ENSG00000271380.1__FPKM:_2000000__name:_RP11-307C12.12__locus:_chr1:154934300-154935099'
'lcl|IsoformID:_ENSG00000222974.1__GeneID:_ENSG00000222974.1__FPKM:_16.4177__name:_RN7SKP228__locus:_chr7:12916308-12916571'
'lcl|IsoformID:_ENST00000476421.2__GeneID:_ENSG00000239708.2__FPKM:_25.0129__name:_RN7SL782P__locus:_chr5:107070475-107070767'
'lcl|IsoformID:_ENSG00000264462.1__GeneID:_ENSG00000264462.1__FPKM:_417.359__name:_MIR3648__locus:_chr21:9825831-9826011'
'lcl|IsoformID:_ENST00000395067.2__GeneID:_ENSG00000066697.10__FPKM:_32.315__name:_MSANTD3__locus:_chr9:103189437-103213511'
'lcl|IsoformID:_ENSG00000239202.2__GeneID:_ENSG00000239202.2__FPKM:_49.0894__name:_RN7SL499P__locus:_chr2:232510710-232510984'
'lcl|IsoformID:_ENSG00000265053.1__GeneID:_ENSG00000265053.1__FPKM:_47.9133__name:_RN7SL321P__locus:_chr3:48421323-48421620'
'lcl|IsoformID:_ENST00000501122.2__GeneID:_ENSG00000245532.4__FPKM:_11.739__name:_NEAT1__locus:_chr11:65190268-65213011'
'lcl|IsoformID:_ENST00000368457.2__GeneID:_ENSG00000163348.3__FPKM:_8000000__name:_PYGO2__locus:_chr1:154929501-154934224'
Another New Observation
- in the totRNA, a lot of the short RNAs are still sequenced as highly abundant, despite the fact the total RNA data says its for greater than 200 bp.
- we probably underestimate the abundance of lots of the short RNAs, which may also contribute to erroneous reads.
- Tested this with E2 data. over-represented sequences are not mapping to non-polyA RNAs that are shorter than 200 bp.