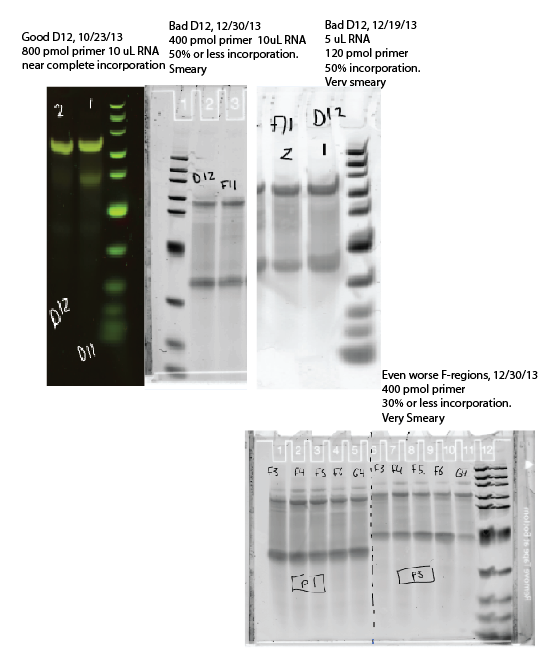
9:45a – 12:15a
STORM analysis
- analyze bead data for BXC double stains
- analyze bead data for PhM Psc
- Finish O/N run of G05
- quick look at ANTC data (just image based drift correction available at present)
- fewer localizations despite bright dyes with MEA. This is expected
- Many clusters not too compact. Radial symmetry of G09/En/BXC generally less evident
ChromatinCropper
Redesigning data storage structures.
- Each saved dot is its own object with its own data
- Each folder is also keeping a structure that has a running record of all the data for all the spots.
- Each spot should get processed individually. We want to look at these one by one and decide whether it’s sufficiently in frame, properly thresholded etc. Not do it in batch and ask if the batch all looks good. This requires a dramatic redesign of step 6 and 7.
Recording spots
- spot per image leaves odd blanks and turned out to be hard to trace back.
- each spot is just going to have all its own image data and all its own meta data saved directly with it.
- just insert save dot in linear array
- potential issue: if you click save dots twice, the data will get double entered.
Step 6: Filter Dots
- Let’s use the dot slider as the advance dot.
- Add a new button to save the dot after examining it. None of this apply a bunch of stuff to all the dots in this image and then chose which ones are good enough to save. Hopefully saving dot by dot also reduces the chance of double entry. It’s no longer a ‘step’ that I might be I’m not sure if I ran it or not. Well, that, and hopefully the printouts to the
Data structures
- large cell arrays of structures not a good format. Hard to index out of.
- i.e. don’t use
dataobject{i}.datatype use dataobject.datatype{i}
New General Purpose functions of interest written today
structOut = IndexStructure(structIn,idx) For structures full of cells and arrays that are all same length (e.g. mlists) and you want to index out of them.clrmap = GetColorMap(colormapName,pts) Returns more colormaps (including all my new black to white via blue, black to white via green etc ones).[stormXZ, stormYZ, stormXY] = List2ImgXYZ(mlist) Generate XZ and YZ projects from 3D data
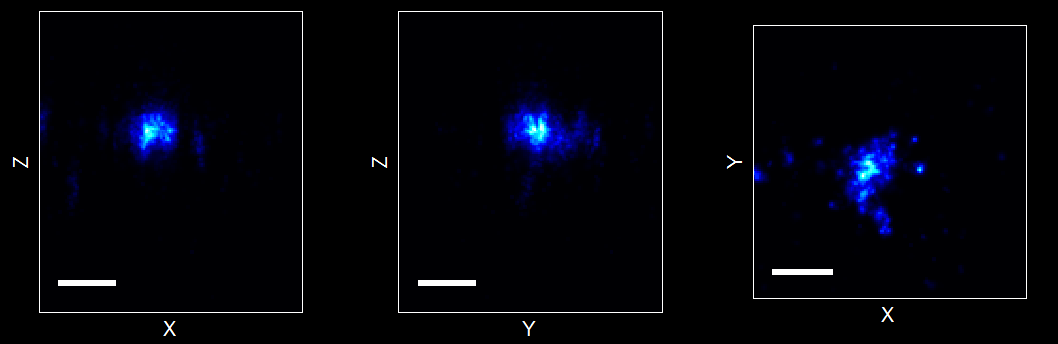
Of interest
git log -p --follow filename the --follow will track the file through moves within the repo and file renaming.
Feedback
- read Carl’s proposal and provide feedback.
- discuss proposal with Carl.
- Still to do!: reread latest version of Hao’s write-up and provide feedback.