9:40a – 1:00a
Goals
- send fig files to Jeff
- Upgrade memory on Cajal.
- Image ANT-C complex
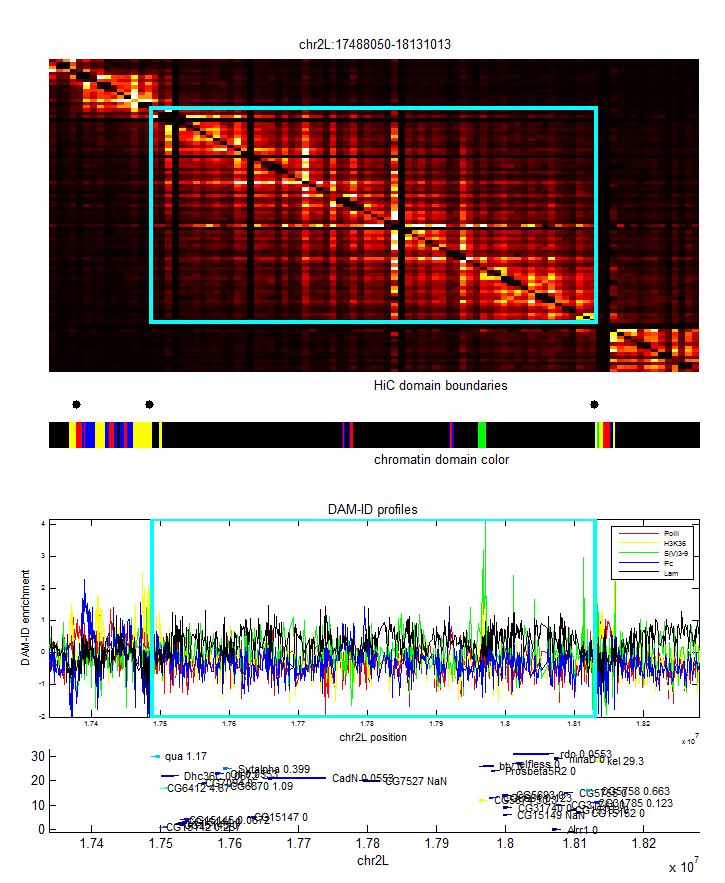
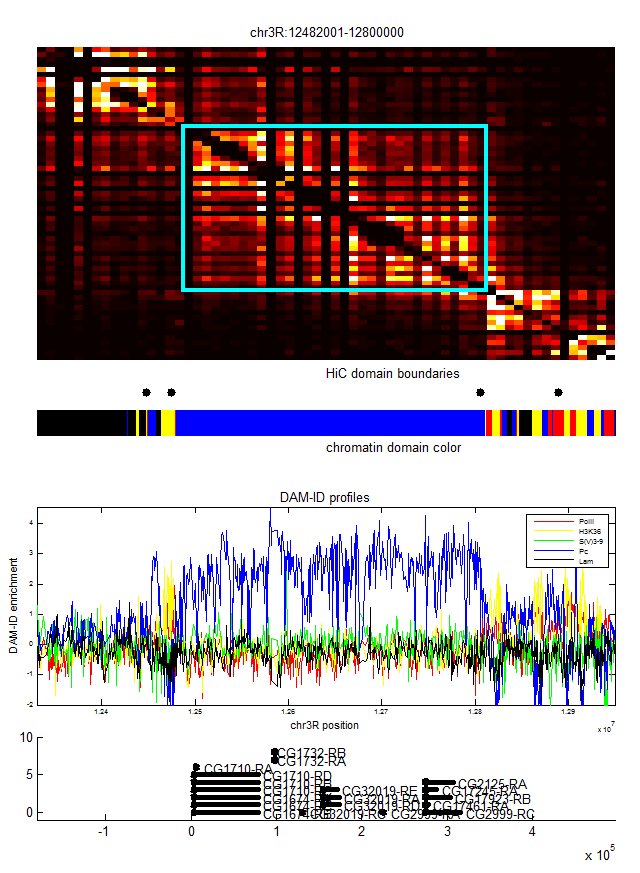
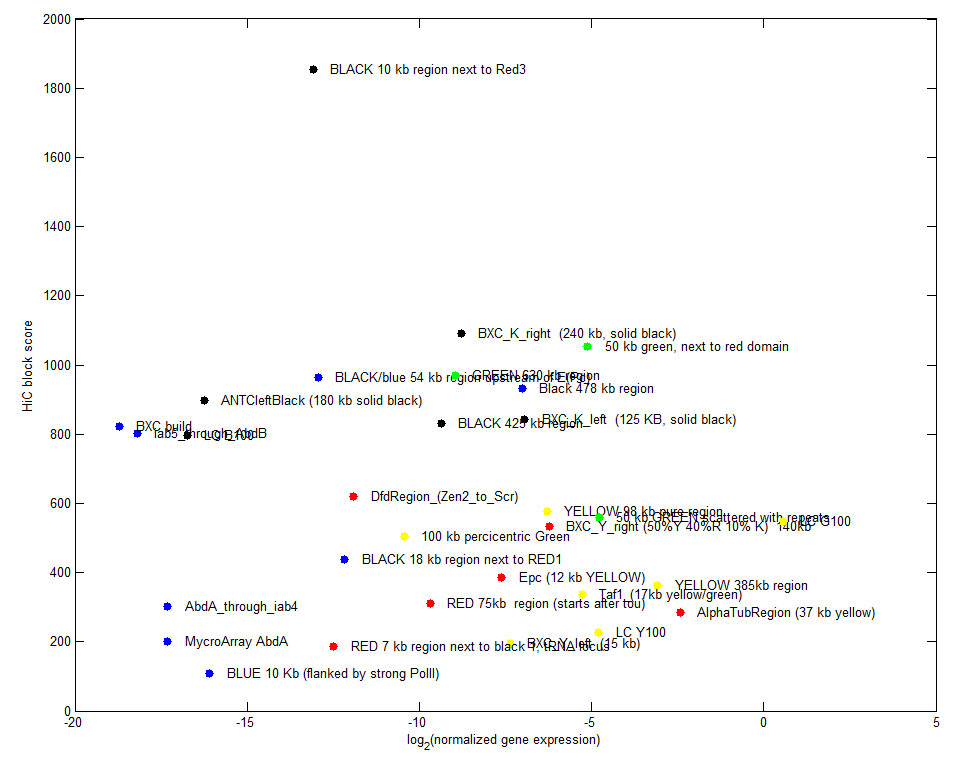
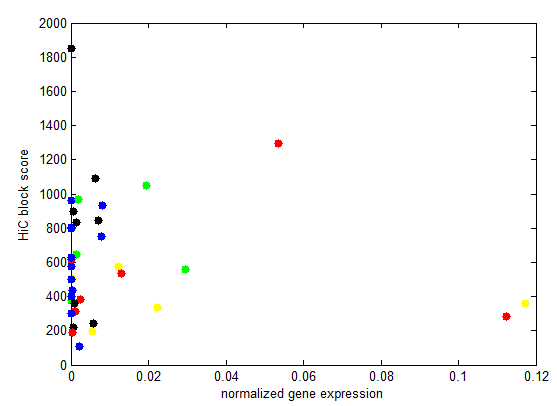
- analyze black K100 (internal black domain)
- plots of internal vs. external.
Lab Meeting
- see notes.
- journal club notes
Computer stuff
- Memory installed on Cajal. Need to upgrade Server edition to use more than 32GB of RAM.
- Windows 2012 Server Standard will do it, and can be upgraded directly from 2008 Server R2 Standard.
- Link to licenses from Newegg for multiple remote desktop for 2012.
matlab-storm development
- weird issue with imagesc not printing indexed data to figure windows seems to have gone away.
- added colormap selection options and tested contrast adjustment in ChromatinCropper
adding colormap selection to STORMrender
- To Do: add drop-down menu next to levels to select color for the channel
Chromatin Project
Presentation
- working on slides for meeting with XZ
STORM
- imaging ANT-C on STORM2
- fresh glox, 2uL MEA 5uL COT, excellent switching (also a brightly stained 400 kb locus).