9:45a – 12:15a
Goals Today
- Rewrite
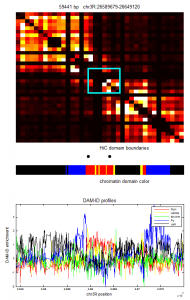
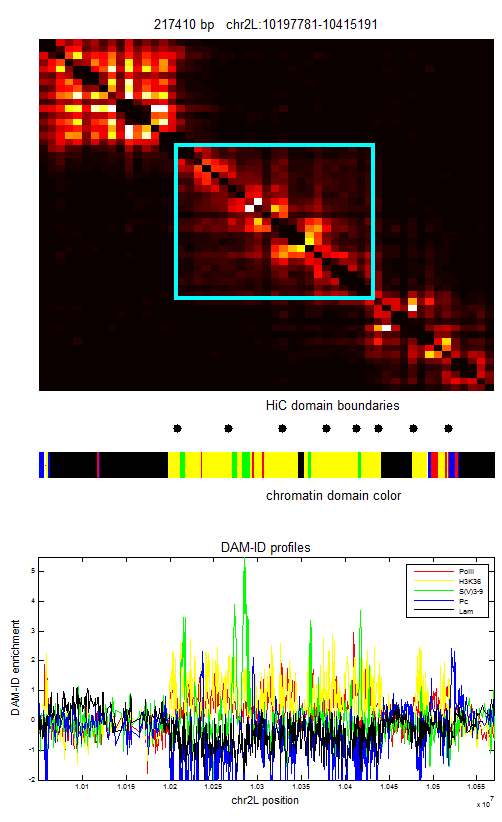
ColorIntervalJumpGap.mFunction to allow different gaps for different chromatin types. - Select new Red regions
Chromatin Project
analysis
- Designing some new regions: notes
- Writing up summary of chromatin regions thus far: see protected notes.
- Let’s try to make a mini-lab meeting like presentation for my meeting with XZ on Tuesday, and plan to order Library 3 between the end of that meeting (if no changes need to be made) and the end of that week (if we change priorities for what goes in there).
- Let’s aim to have all the potential regions folded and sorted with their primers before then.
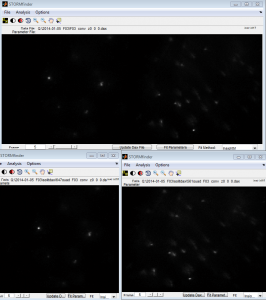
STORM
- imaging F03-647 F04-750.
- calibration image looked pretty good
- 750 background very low, dots pretty faint though / fast bleaching. I’ve gotten much brighter 750 in the past. Might want to increase 750 probe conc. slightly.
- switching not very bright (2.5 uL BME + 5 uL COT). Could use brighter dyes. Maybe try with MEA tomorrow. That’s my only day-time ‘scope time this week. Can try with same coverslip.
Project 2, code organization
- discuss public vs. private reps
- discuss folder organization
Ph Project
- Start analysis of Psc PhM data