Introduction / Motivation
Current library 3 design is still missing some key regions we would like to investigate very soon. For example there is no concerted attempt to bring red regions back into the fold. Preliminary observations of red regions suggest we have some interesting things to say about these, so I think we should really keep them in the survey as their own thing.
Selecting more regions for Library 3 to fill in gaps:
New candidate red regions
- chr2L:12423661-12550000 length: 126339.
- chr3R:18981944-19055219 length: 73275
- chr3R:19105077-19172133 length: 67056
- chr2L:19134769-19179775 length: 45006
- chr3L:8651129-8702767 length: 51638
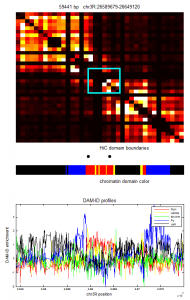
- chr3R:26589679-26649120 length: 59441 — Nice Red w/ H3K36me3 and blue/black bounds.
- chr3R:9876027-9898927 length: 22900 – Red/Blue
- chrX:12622514-12649591 length: 27077
- chr2R:8146036-8162625 length: 16589
New Candidate large yellow
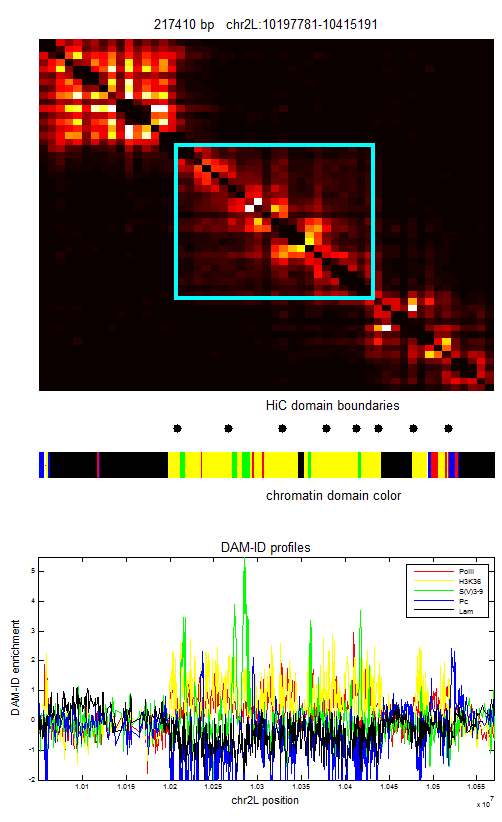
- chr2L:10197781-10415191 length: 217410 — reasonably solid
- chr2R:19691887-19968531 length: 276644
- chrX:15590302-15841088 length: 250786
- chr3L:9344016-9482324 length: 138308
Some red and yellow regions of interest:
Relevant Code:
- see
FigureCode\Lib3RedSilentFlanks...
Green
- intermediate regions don’t really exist, my current ones are mostly pericentric regions split on tiny red/yellow boundaries
- some interesting variation among the miniscule, tiny, wee, and small sizes (~2 kb, 10 kb, 15 kb, 20 kb), but I don’t want to pursue that now. (see notes below on combo regions)
Interesting combo regions
- Su(var)3-9 +/- H3K36me3 (green)
- Su(var)3-9 occupied genes (in otherwise euchromatic regions). Often have distinct many-exon structure
- Su(var)3-9 un-occupied genes embedded in deep Su(var)3-9 heterochromatic (percentric) regions.
- PolII +/- Pc (red)
Screen library 3 properties
- goal: A bunch of the regions not selected specifically for domain color size combos still fill useful positions on this matrix.
- Try out new approach of keeping all library data increasingly in matlab structures instead of raw text and flags to parse from fasta headers. That’s a fine end step.
Organize library 2 in alternate grouping than color
- by HiC domain scores (come up with some way of quantifying this)
- # of domain boundaries
- box score — give points for off-diaganol mass, linearly normalized sum of HiC region
- by DAM-ID tracks more directly.
- Combos: e.g. H3K36me3 high and Su(var)3-9 high.
- might be a subsorting
- by GC content? probably no good.
- By gene density?
- would be good to integrate a gene display into matlab region viewer. This may take some work.