September 2025 M T W T F S S « Aug 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 Categories
- AP patterning (13)
- Blog (1)
- Chromatin (88)
- Conference Notes (72)
- Fly Work (54)
- General STORM (25)
- Genomics (134)
- Journal Club (22)
- Lab Meeting (66)
- Microscopy (79)
- Notes (1)
- probe and plasmid building (58)
- Project Meeting (3)
- Protocols (13)
- Research Planning (74)
- Seminars (21)
- Shadow Enhancers (59)
- snail patterning (40)
- Software Development (5)
- Summaries (1,412)
- Teaching (9)
- Transcription Modeling (40)
- Uncategorized (10)
- Web development (19)
Links
Tags
analysis cell culture cell labeling chromatin cloning coding communication confocal data analysis embryo collection embryo labeling figures fly work genomics hb image analysis image processing images in situs Library2 literature making antibodies matlab-storm meetings modeling MP12 mRNA counting Ph planning presentation probe making project 2 project2 result results sectioning section staining shadow enhancers sna snail staining STORM STORM analysis troubleshooting writing-
GitHub Projects
Wednesday 01/08/14
10:00a – 5:30p, 9:30p – 11:30p
Ph project
- Ph project model figures for Ajaz. See post
- Reorganized figure producing code (see
Code\Results)
Chromatin analysis
- copying F05 data over to Probox 7
- Running analysis of F03 and F04 while copying data to same disk not good for drive communication speed. makes it very hard to analyze data off the same disk at this same time.
- this is in fact a known bad idea. Cancelling data copying.
Library design
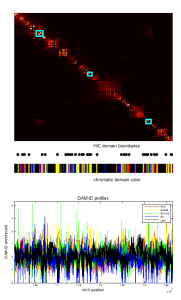
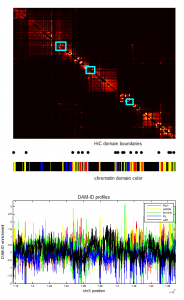
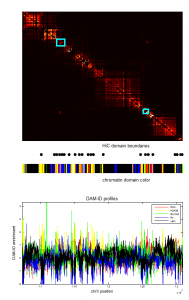
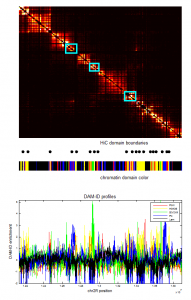
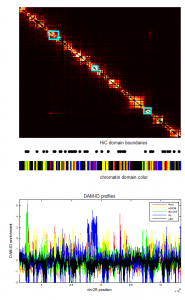
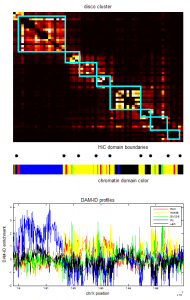
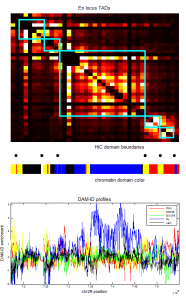
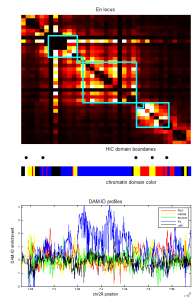
- screening HiC data for off diagonal contacts.
- need to go back and choose a region inbetween as control comparison.
- region selection maps
STORM
- Overnight imaging of BX-C composite (F03 + F04 + F05 P1s).
- clusters look pretty good.
- focus lock having more serious issues as several positions on this coverslip. Will probably lose focus at some point in the night.
Discussions
- discuss probe design with Sebastian for yeast.
- Will help generate probe sets tomorrow
Tuesday 01/07/14
10:00a – 10:15p
chromatin data collection
- Finish O/N STORM on F04.
- Transfer data from F03 and F04 to Monet
- start analysis of F03 and F04 data
- wash out stains for new BX-C regions: F05, G04 (ANTC), F03-F04-F05 combo, F05-A647 F06-A750.
- image F05 regions (hope these work!)
Ph project.
- fix error in list_pars.txt files for PhM 750 data.
- crop cells from PhM
Data for Ajaz
- Working on new model figure as slides for Ajaz — still haven’t finished this!
- New Pc clustering — existing data pipeline for analyzing this is pretty good. Moved functional form of script into a Results code folder.
Ph project observations from new data set
- Substantial over expression — by quantifying localizations its 5x the wt levels. 100K localizations in both 750 and 647 per cell, whereas cells with no nuclear enrichment of 750 flag have 20K localizations of endogenous Ph.
- issues with z-focus
- wt data was taken at lock target 0 for both 647 and 750. This seems not to align focuses– substantial new molecule regions evident in each channel not evident in other channel. Spatial distribution through nucleus seems to suggest a z issue. For example, the interior of the nucleus will be hollow and one channel will show a more filled middle and the other a more extended periphery.
- M data was taken at lock target 0 for 750, +60 for 647.
- wt and mutant data good cluster overlap
- images align better with drift correction turned off. Imaged based drift correction is just creating trouble.
- caught and fixed a few little bugs in STORMrender
Mentoring
- project discussions with Bogdan, 11-3p
Project 2: error checking
- check probe alignment
- wrote short script to BLAST against new internal database
- legacy BLAST returns no hits. is it legacy BLAST that is strand specific not oligoArray? Or maybe I don’t have it configured right
- BLAST+ works great, sequences all check out as desired. Only matlab readblastlocal doesn’t succesfully import the strand (should be Hits.HSPs.Strand, but this field doesn’t get created, even though I can see it on the Blast output).
- updated my matlab BLAST function to default to display the name and sequence alignment of the top hit. would be awesome to get strand as well.
Monday 01/06/13
10:00a – 11:30p
Meetings
- lab meeting (post-doc interview). See notes
- post-doc chat
- fixing up slides for post-doc chats, moving onto google docs so that the presentation is mobile
- google docs having issues loading images in presentations that used to display just fine.
- a number of images display properly when run in presentation mode. some still fail to load.
- libreOffice keeps freezing when I try to update my current set of slides. what is this?
- attempting to download. This actually works and displays figures correctly
- notes for next time: fewer stories, more background.
- post-doc dinner
Chromatin imaging (single color)
- Finish O/N STORM of F03 spots on STORM4.
- Set up new O/N STORM of F04 spots on STORM4
- test out Hal upgrades on STORM4?
cell culture
- Set up new 12-well plate of Kc167 cells for fixation and hybe prep
- passage cells (new media to both active flasks).
- fixing cells (settled out perfectly confluent).
New stains
- G4-P1-A647
- F5-P1-A647
- F3 + F4 + F5 -P1-A647
- F5-P1-A647 + F6-P3-A750
Ph project
Modeling
- reasonable results for PhM effects in capping model
- need to re-run for diff binding model (now in process)
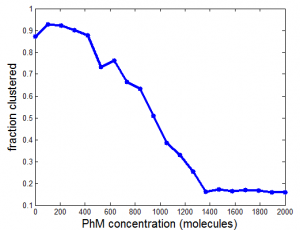
Data analysis
- start processing cells through STORMrender (used to parse out individual cells) for Ph analysis.
- over-expression I think is killing the structure. The lower expressing wt look much better.
Protected: post-doc interview: 01/06/13
Posted in Lab Meeting
Comments Off on Protected: post-doc interview: 01/06/13
Sunday 01/05/2014
10:30a – 8:00p, 10:00p-11:50p
literature
- continue reading Berlivet et al PLoS Genetics on HoxA
- ID 19 candidate enhancers by ChIP and 5C
- 8 of 10 tested LacZ transgenics show limb bud enhancer activity
TAD contacts in silent tissue
- Shh signalling blocks processing of Gli TF into its truncated repressor form. Consequently GliR only exists in the anterior domain of the limb-bud in wt. In Shh nulls, GliR is everywhere.
- In this condition enhancers with GliR binding sites (e.g. e5) are silenced (no longer drive LacZ transgene expression). 3C contacts are still detected, though not as strong as wt. (This could mean the true interaction frequency is unaltered, and the promoter, not being transcribed, forms fewer contacts. Or it could be the authors interpretation that interactions are preserved yet weaker).
- 5C analysis in Shh nulls recapitulates wt contact pattern.
coding matlab-storm: STORMrender
- switched to a STORMrenderBreakOut branch to export STORMrender functions
- Completely exported all subfunctions from STORMrender
guidatacommands run fine from external functions, just need to passhObjectas well- No need to return
handlesand callguidatawithin the main GUI function (though I do this a good bit at first) - Now I need to see if how many of these are ready to use as crafted by my CC GUI. Wasn’t thinking of using by multiple GUIs…
- In the short run the Break out should make the details of changes much easier to track. If Bogdan and I get up to sync this way with CC it will be much smoother. All going to take some more work though.
more coding goals: bead-data compression
- Really should get back to bead-data compression soon, running out of disk space quickly at present.
- I recommend recording the compression in the new info file, and checking the infofile for compression when reading in the dax. Otherwise we could record it in the list_pars file, but since it’s a dax modification it really should go in the dax header file, i.e. the .inf file.
Chromatin staining
- F3 sample finally stains! (there was no good reason this one should have failed twice in the first place, this is nice confirmation.
- Let’s get some more BX-C sub-cluster imaging.
- Should also plan to stain
- Should keep the experimental front busy until I can get back on the 750 for multi-color
- Sonicating new batch of 561 beads. Making new batch of Glox.
Chromatin analysis
- F12 stains pretty backgroundy in all images checked so far. Need to add more filters.
- G1 G2 looks more promising.
Saturday 01/04/13
10:00a – 11:55p
(sailing canceled do to malfunction of committee boats)
coding matlab-storm
- RunDotFinder “skip existing” option on DaoSTORM I believe is interpreted as resume and in insightM as actually skip. This should be double checked and made clearer. Maybe we want a legitimate skip as well to be an option for DaoSTORM.
- Fixed up alternate binname functions (can pass it
'DAX'to include the current daxfile name and append or prefix it with a new id. Can also pass it'#'to insert image index (based on results of dir daxroot).
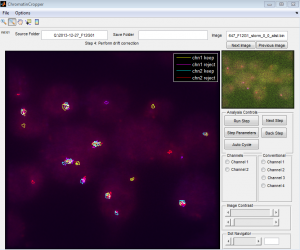
Mostly writing new multi-region version of ChromatinCropper
- Not all movies have bead data analyzed yet (I suppose I was planning to downsample all these files. I should probably set that up to run soon).
- note with no averaging and single round, daoSTORM seems to out perform insightM on bead-data
important observations in drift correction
- smoothing may work alright, but has serious issues at the boundaries. We use these boundary points to correct drift between movies, so this is an issue.
- we could compute a warp based on the bead images.
- fixed NaNs is
fastsmooth.m(rather than usinginpaint_nans.m, both from Matlab file server). Original author had tried to make it NaN friendly but got foiled by the+and-operators which still return NaN. (usenansum([a, -b])rather thana - bto subtract and ignore NaNs). - also switched to narrowed averaging window rather than just zero padding of ends during smoothing.
- still need to do something about vibrating beads.
STORM analysis
- Bead data from F12G1 not analyzed. Processing now on Cajal
- no data from G1G2 analyzed yet. Processing now on Cajal
- G1G2 data not nearly as bright as other data sets. Should collect more data with a fresh buffer when we get 750 time again.
- G1G2 slide is pretty low density on beads — lots of images have none.
Friday 01/03/2014
remotely 10:30a – 11:30a (snowstorm)
Winterize bike: 11:30a – 2:00p
lab: 2:00p – 9:00p
Goals
- switch road tires for mountains for better stability in the snow
- Design new primer collection for chromatin library 3.
- Pick more regions for multi-color experiments: split up BX-C again.
- finish new year’s letter. update contacts list. send letter.
Library 3 preparation
- Re-dissecting BX-C. Using embryo CTCF sites for boundaries.
coding: ChromatinCropper
- started new
developmentbranch - splitting all the functions and all the RunSteps out of the main GUI into separate scripts in separate folders
- fixed some more bugs in ReadDax. Should have switched back to
masterto do this. - merged
developmentintomaster. branchalistairstill has the original ChromatinCropper, and master doesn’t really care what happens to ChromatinCropper at this point anyway since it’s still in the Beta folder. - switched back to
developmentto keep working on ChromatinCropper. - clearly need to start saving my conventional images as conv_z0 to indicate the starting plane.
Thursday 01/02/2014
9:30a – 9:00p
Ph project
- Finish O/N STORM run
- Take IR bead images (slide is also getting clumpy. Maybe we need to stick these down with EDAC. I think making slides for long term storage with Glox is also a bad idea, brighter to start with but shorter shelf-life).
- start copying data
- PhM data still fitting
- Psc Ph data finished analyzing (72 hrs).
- start re-running Ph folding sims with revised correction of clustering based on unbound fraction.
Variable density of Ph staining (all cy7-Ph-flag staining in Ph Wt background):
More drift analysis
Installed inpaint_nans.m from matlab file exchange to interpolate through NaNs (frames in which beads were not detected). This is required for the cross-correlation to work properly.
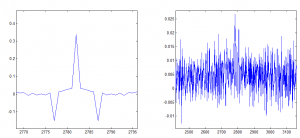
Averaging 20 frames (5 Hz), and then smoothing 20 frames, cross-correlation between beads on small time frame is mostly correlated
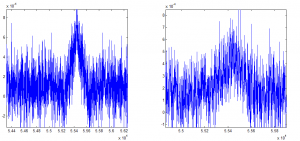
Auto-correlation is shown on the left, cross-correlation to other beads is shown on the right.
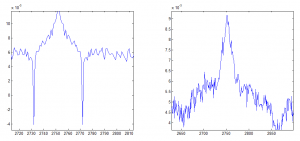
Determining ideal down-sampling
- comparing 20 frame averaging to 400 frame averaging
- 20 frame averaging + 20 frame smoothing filter looks better, let’s see how this compares to 400 frame averaging.
Library3 Prep
New cell staining
- G1-P1 (positive control)
- F3-P1 test
- F4 P1 test
- used 12/14 fixed and prepped cells.
- No RNase treatment (these are all fully silent regions).
- .5 uL secondary, 2 uL primary each. Used previously successful secondary aliquot (I had doubts about some of the aliquots, switching aliquots earlier correlated with success / failure of staining). Used hybe dilution mix.
- pre-hybed about 5 min at 47C prior to adding probe, then about 1 hour at 47C after adding probe while heat block was stabilizing for denaturation.