9:30 am – 7:00 pm, 9:00 pm – 11:00 pm
Deep seq prep
- continued with deep-seq prep
- finished second strand synthesis
- ran clean up on DNA-XP beads from Bauer
- ran adaptor ligation. premixed adaptor and enzyme mix (2 min on ice), this is bad it will increase adaptor dimer.
- we’ll quantify this on the tape-station and see how much trouble it will cause.
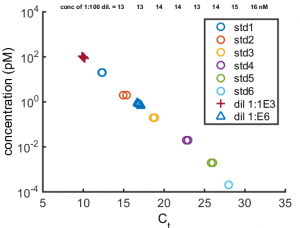
RNAi quantification by qPCR
- set up QPCR plate, same organization as last time.
- cDNA row order: 4W, 4P, 2-4W, 2-4P
- ran tape-station (only 4 lanes left, ran ladder and lanes 1-3). looks decent.
- ran kapa quantification
seq prep
- previously used 1,2,4
- setup 1 & 2 as repeats with 6,7,8 as multiplexed. (Illumina 1,2,6,7,8)
- run new 1-5 multiplexed together (Illumina 1,2,3,4,5)
- submit for 20 uL, 2x single lanes.
References to cut (sad, I like these papers)
- Killian 2015
- Kharchenko 2011
- Ernst 2011 (ENCODE)
- Ram 2010 (ENCODE)
- Li 2015
- Galupa 2015, (Heard Lab)
- Rivera 2013 (Ren Lab)
- Gomez-Diaz (Corces lab)
- Rosa 2014
maybe cut
- Gorkin (Ren Lab)
- Gibcus (Dekker Lab)
- Bernstein & Lander 2007 (~idea founder but old)
- Simon and Kingston 2013
- Boyle, Yamada, and Beliveau 2012 to supplement, Chen 2015 to supplement?
notes
- Bickmore 2013;Gibcus 2013;Levine 2014;Gorkin 2014;Sexton 2015;Galupa 2015;
- 8-10 Bernstein 2007;Rivera 2013;Bickmore 2013;
- Bickmore 2013;Gibcus 2013;Levine 2014;Gorkin 2014;Sexton 2015;Galupa 2015;Bickmore 2013 (1);
