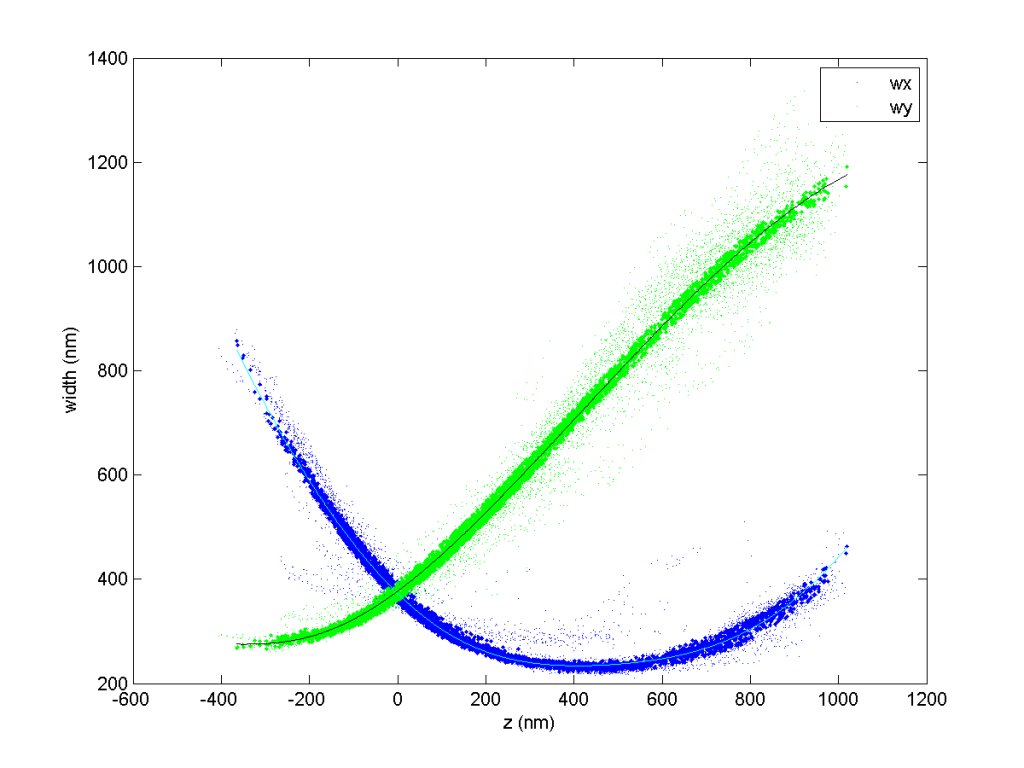
with single iteration, Hazen 3D-DaoSTORM avoids splitting molecules more distant from frame
A few notes on 3D-DaoSTORM
- First 4 iterations are computed with 4x the input threshold. So if using less than 4 iterations you want to quarter the threshold to get the same results.
- 3D-DaoSTORM iterative subtracts the fitted pointspread function (with whatever elliptical fit it computed).
- 3D-DaoSTORM produces separate ‘averaged-molecule lists’,
*_alist.binand raw molecule lists*_mlist.bin