11:15a – 11:45p
Ph Project
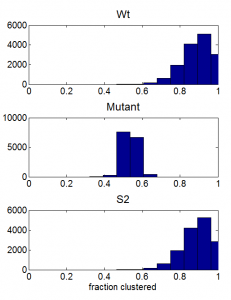
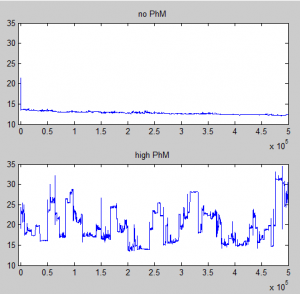
- Highly variable radius of gyration measurements for case with binding — suggests not equilbirated. Binding strength probably much to high. Up to .90, run again. Also increased density of K27me3 sites from 50% to 80%, Hoping to reduce a little bit the run-to-run variation in topology, which is extreme in terms of Rg (Mutant bounces around 15 to 35, some Wt get locked at 10, some at 15, some at 25, only very gradual movement).
- relaunched simulations for +/- K27 constraint and Capping vs. Diff binding for PhM concentration, using
Ebond = 0.9(may still be too low), and 80% K27me3. - Added new subsampling routine to Ph clustering to get better distribution stats (i.e. bootstrap to estimate variation)
- PhM variation in fraction clustered is still much lower than in measurements. This is as expected, because other measurements suggest the large variation in phM clustering is due to variation in PhM concentration, and we keep this fixed in the current simulations. Maybe I should run another set of simulations sampling PhM from a broad range of concentrations?
PH STORM
- Testing stains on STORM2
- cells didn’t stain in 750
- Either cells didn’t transfect, didn’t induce, didn’t label primary or cy7 secondary / dye is failing us again
- Check control cells — PhM labeled with m anti-flag and 647 anti-m. Mostly unstained as well. found 1 well stained example. Seems to be a transfection or induction problem.
- Check Wt cells tomorrow
Cell staining / bead test
- G03 P1 1.5 uL, 1 uL S1
- new EDAC, freshly dissolved in MES
- vs. old EDAC
Oligo Secondaries
- Put together draft figure of oligo secondaries
- running more of the oligo secondary data with ChromatinCropper
- set up new folder in Projects of OligoSecondaries (used to be a component part of the Chromatin project)
- observed interested loop domains, should do some follow up on this
- finished draft figure
- ran KS-test (kstest2) stats on cdfs,
- sent draft with comments to Brian
Testing multi-color
- On STORM2
- dots in neither channel looks strong: either the G5-G6 or the G1-G2.
- G1 G2 has more convincing pairs. Really want to get this G1 647 at least to be much brighter than this. Maybe its’ a probe concentration issue? Or something rogue went wrong with the stain / cells?
- We’ll see how the G03’s look tomorrow from the bead test. Then maybe I’ll try increasing the probe concentration. After that it’s back to probe synthesis and hope the next batch works out better.
Discussions / advantage of chromatin STORM
- small region inside a large region — conventional domains have no boundaries
- vs two small regions separated by notable distance
- or not so small regions with dense sides near each other
- Also illustrate the contrast effect — such as a small projection from a dense bright domain — even if larger than a diffraction limited spot, the contrast difference makes this project go undetected — it’s still in the scattering zone of the bright domain.