9:30 am – 9:15 pm
MERFISH
- working on aligning data
- current pentanotch no quadview approach requires different approach to beads
- try multilabeled beads (homemade tetraspec with 750 dyes)
- try using labeled cells
Chromatin meeting with XZ
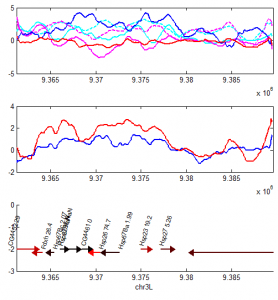
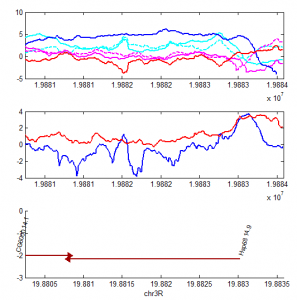
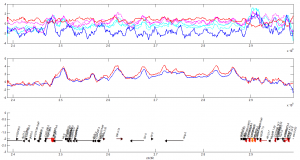
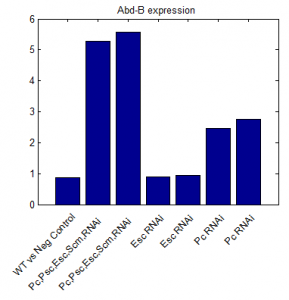
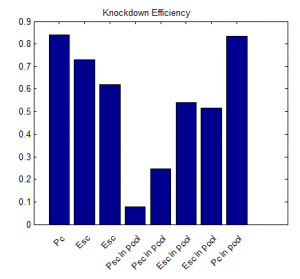
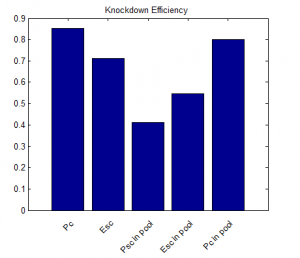
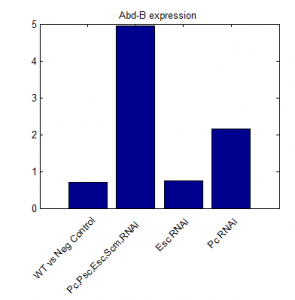
- priority on PcG knockdown
- use immunostain of protein to quantify protein efficiency KD
- try to increase KD efficiency of individual KDs.
- keep focus on multi component KD for imaging work
Apps and Conferences
- finalize DF files, submit to Matt
- send signature form for internal signatures
- working on slides for SDB meeting
- getting a few things for ML party at SDB
To order
- secondary sequences fused to common sequence
- more RNAi primers
- more PCR primers