12:00am -7:00am, 11:30am-7:00pm,
Manuscript
- finished submission, ~7:00 am. yay!
New material to work with:
Embryos
- flipped fly plate, 12:30 pm.
- plan: 3 hr collection + 2 hr age -> 5:30 pm
- flipped cage again 3:30 pm
- depolymerizing formaldhyde at 70C in incubator (should order more, almost out). Need more heptane too.
- plan: fix embryos
- fixed embryos at 5:30 (2-5 hr collection). Good yield. With red label, marked with 2-5hr yw and 15-06-13 in methanol in -20.
Cells
- started new Kc167 culture going from frozen stock, 1:00 pm
- passaged to remove residual freezing media.
- density is excellent, could move to large plates tonight / tomorrow.
Additional prep
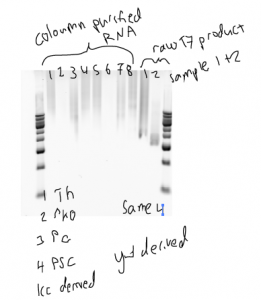
- SN points out embryonic derived PSC Su(Z)2 Pirrotta and Schwartz labs
- see experiment planning post describing plans, reagents and resources.
Ordered
- Heptane
- ultapure formaldehyde
- Sneider’s media (for the RNAi and new cell lines. at least necessary for the serum shock transfection).
MERFISH
- catching up on the recent progress be reading the team lab-notebook entries I missed.
Paperwork
- finish descriptions of patents for DR progress report.
- sent forms for signature to XZ.