9:40 am – 5:00 pm
Goals & progress
- prep and stain embryos with new L7 library. -> DONE
- order food for group meeting. -> DONE.
- pick up suit for interviews. -> DONE.
- work on equipment lists. -> Extracted parts list for STORM5
- get estimates of microscope room specs. -> contacted Mike Paterno
- contact UCSF about dates -> Scheduled: Mar 4th and 5th. (Thur + Fri).
- order Alexa 647 labeled probe complimentary to L7 (thought I did this earlier but check records today and not ordered).
Experiments
- L7 library staining embryos (3 uL library to 25 uL hybe dilution buffer). Stained 2 coverslips.
- denatured with block set to 90C. Block reading 78-80C during incubation once cold glass slides and water are added. Thermometer reading 85C.
Image results
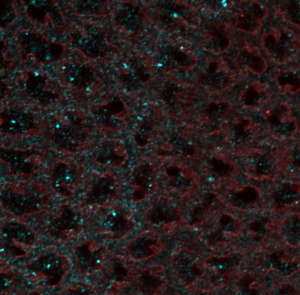
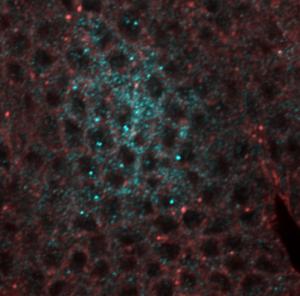
- RNA probes clearly labeled better than smFISH DNA probes for confocal imaging.
smFISH hb-647
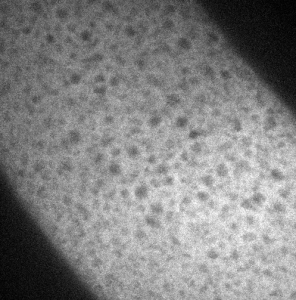
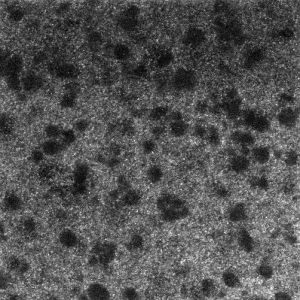
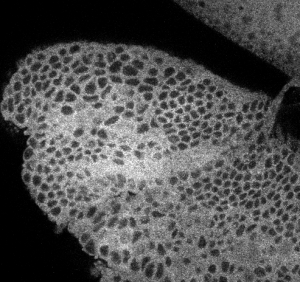
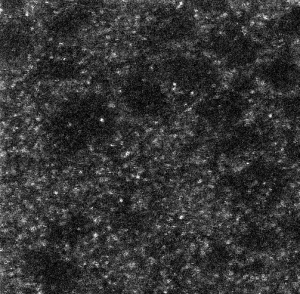
smFISH RNA probes with antibody
Code
- start writing (and streamlining) code for an engrailed locus library.
maintenance stuff
- started new box of common MERFISH supplies for team
- started new excel sheet for logging my lab purchases
- created new excel sheet indexing all parts purchased for STORM5