November 2025 M T W T F S S « Aug 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 Categories
- AP patterning (13)
- Blog (1)
- Chromatin (88)
- Conference Notes (72)
- Fly Work (54)
- General STORM (25)
- Genomics (134)
- Journal Club (22)
- Lab Meeting (66)
- Microscopy (79)
- Notes (1)
- probe and plasmid building (58)
- Project Meeting (3)
- Protocols (13)
- Research Planning (74)
- Seminars (21)
- Shadow Enhancers (59)
- snail patterning (40)
- Software Development (5)
- Summaries (1,412)
- Teaching (9)
- Transcription Modeling (40)
- Uncategorized (10)
- Web development (19)
Links
Tags
analysis cell culture cell labeling chromatin cloning coding communication confocal data analysis embryo collection embryo labeling figures fly work genomics hb image analysis image processing images in situs Library2 literature making antibodies matlab-storm meetings modeling MP12 mRNA counting Ph planning presentation probe making project 2 project2 result results sectioning section staining shadow enhancers sna snail staining STORM STORM analysis troubleshooting writing-
GitHub Projects
Protected: project 2 update 07/08/14
Posted in Summaries
Comments Off on Protected: project 2 update 07/08/14
Tuesday 07/08/14
10:00 am – 8:00 pm, 11:00 pm – 11:50 pm
Sequential staining
- F06 at least stains this time, some spots quite clear. However staining not very good and STORM pretty weak.
- Tried flowing more secondary for an extra 2 hours and washing without formamide.
- stain not much improved.
- attempting STORM.
- STORM imaging of previously imaged regions looks horrible.
- maybe this F06-p1 probe is not good. Test it independently tonight? swap the order of the stain?
Data analysis
- launched analysis for
- L3C02,
- L3E05 and
- L3D8to12
Troubleshooting continues
- restain, this time apply S1 O/N. Should answer the question does the F06-P1 probe work. Also added earlier batch of F06-P1
- also stained each batch of F06-P1 + S1 independently
- test F06 stain tomorrow morning. If this works I bet we can get BXC to work second (fingers crossed).
Additional new stains
- D12-P1 (2 uL) + S1 (.5 uL)
- D11-P1 (2 uL) + S1 (.5 uL)
Project 2
- see notes
- screwed some things up in probe making.
Oligo Secondaries
- playing with scalebars
- made all images square
- Need final review of Fig 2 for Brian
- Need additional images for Supp Fig 14 for Brian.
The other things
Literature
- reading Helin lab paper on PRC2. Very interesting.
- proposes PRC2 purely maintenance role, recruited to CpG islands that lack dense nucleosome arrays.
Department mail
- finally checked this (to pick up rubber cement).
- one of my IDT shipments went to department mail?! (extra common-primer)
- Thesis gift from AMOLF
- nice card from Arnosti group.
Monday 07/07/14
9:00 am – 5:00 pm, 6:00 pm – 11:00 pm
Meetings
- project 2, team meeting, see email
- lab meeting, see notes
- journal club, see notes
Sequential staining
- imaging BX-C. Looks bright and clear again.
- try competing out S3 probe with unlabeled S3 at ~30C (stage still heating). — no obvious short-term effect
- try competing out S3 probe with unlabeled S3 at 37C O/N
Oligo-secondaries
- tweaking figures
Fly work!
- flipped fly stocks.
- One of the Esc stocks definitely dead.
Posted in Summaries
Comments Off on Monday 07/07/14
Journal club: whole animal light-field microscopy
(Prevedel et al Nature Methods 2014)
Light-field microscopy –
- capture 3D strcuture of object in single snapshot
- first developed in 2006
- array of micro-lenses, different parts of sensor get different z-fields
- going deep spread out image, lower signal to noise, sacrifice SNR /x-y resolution for time resolution
Light field deconvolution microscopy
- goal: use deconvolution to improve the x-y resolution that gets compormised by the low SNR
- put beads into agarose, measure resolution on scale of 1 um (little worse) (?)
- calcium imaging (chemical indicators or genetically encoded indicators (Preicma, GCaMP).
- Ca dyes best for change in Ca conc, not so good to measure aboslute levels.
- this paper uses GCaMP.
- image whole worm, 30 um deep.
- don’t actually separate neighboring cells. But do get individual cells resolved over substantial depth.
Posted in Journal Club
Comments Off on Journal club: whole animal light-field microscopy
Protected: Group meeting 07/07/14
Posted in Lab Meeting
Comments Off on Protected: Group meeting 07/07/14
Sunday, 07/06/14
10:15 am – 4:00 pm, 10:30 pm – 12:00 am
Imaging today
single color
- finished imaging Do8toD12
- started data transfer
- started imaging C02. Too many spots per cell in many cells. This is not good, something weird with this locus (or probeset), might be best to skip it for now.
- took a conv mosaic and some STORM images to document these observations in C02.
- set up new scan of E05
Sequential stain troubleshooting
- went to set up new sequential stain, found previous probes F03 to F05 p3, and F06-p3 not P1.
- I suspect this is the reason I could not get P1 to stain
- also means can’t trust the BX-C imaging data from the last few days. Too bad, have to repeat all that.
- set up new stains
- F03 to F05-p3, .75 uL each + S3 1.3 uL, + F06-p1, 1.5 uL
Oligo secondaries
- finish writing methods section for simulations
- sent this and some minor tweaks / revisions to Ting et al.
Project 2 probes
- set up T7 reactions
- started reactions without RNasin. Added at 20min.
Posted in Summaries
Comments Off on Sunday, 07/06/14
Saturday 07/05/14
10:30 am – 9:00 pm
STORM imaging today
Sequential hybes
- repeat staining of F06 not much improved on BXC-F06 try 2. Hardly see any stain (just background?) Attempting to image these anyway.
Imaging new stain: D08toD12
- stains look great. Switching looks great.
Reading
Project 2
- start time 1:45 pm.
- re-clean and paper RNase free zone upstairs
- making probes.
- E1 8 wells A01 + A02 — combine and use 25 ug columns
- E5 8 wells A09 + A10
- E25 1 well E01 + E02
- E26 1 well E03 + E04
- E27 1 well E05 + E06
- E28 1 well E07 + E08
- E29 1 well E09 + E10
- E30 1 well E11 + E12
- Make dilution of primer plates.
- Stock concentration = 200 uM
- need 1:40 dilution. Do 4 in 156.
- Master mix
- 25 uL Hot-start Phusion
- 5 uL 1:100 library
- 5 uL 5 uM
- In parallel, dilute my primers 4 in 78 (10 uM working concentration)
Oligo secondaries
- working on methods description for simulations.
Data analysis
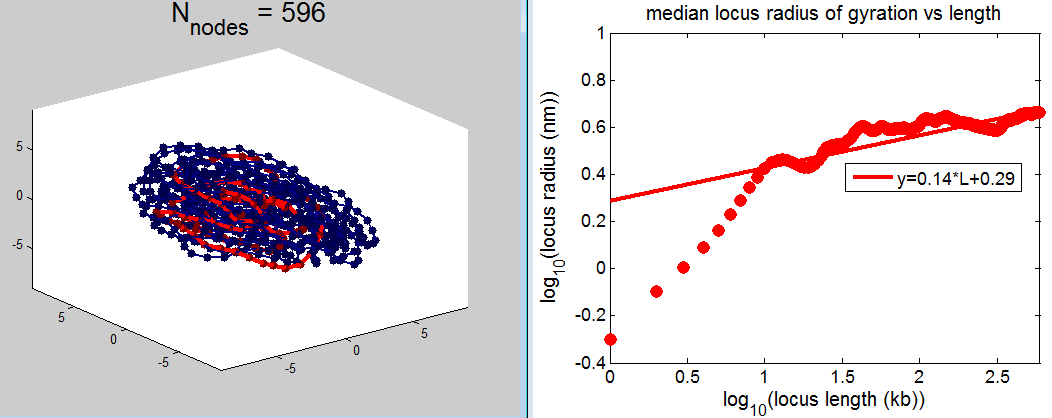
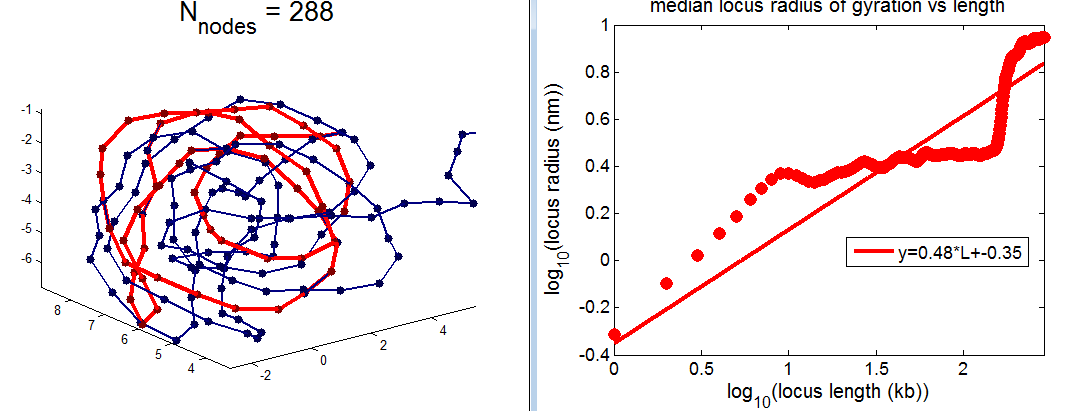
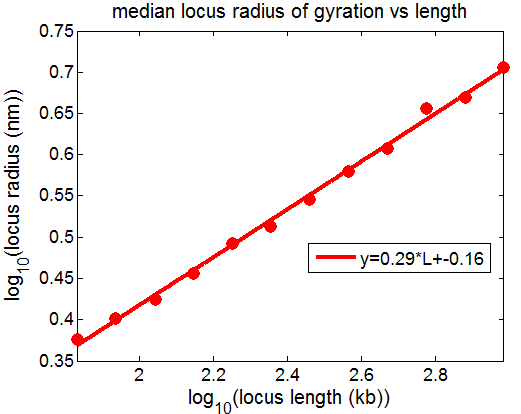
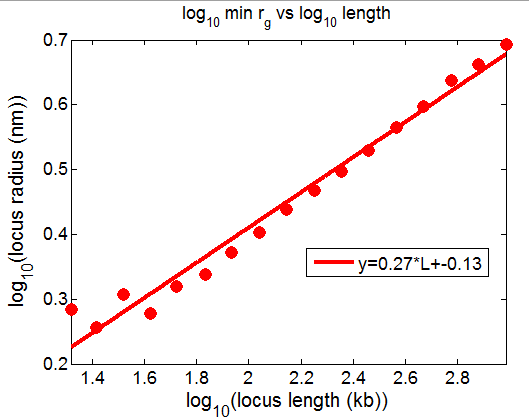
- picking out regions of yellow chromatin to illustrate scaling
- launched spot fitting data on recent BXC attempts.
Posted in Summaries
Comments Off on Saturday 07/05/14
Friday, July 4th, 2014
10:00 am – 1:10 am
Discussion
- sent feedback to Ting about proposed revisions for fig2
- sent comments to Hao about presentation for this evening
- project 2 team meeting
- meeting with XZ discuss project 2 partitioning (see emails).
Analyzing STORM data
- C10 Analyzed through image 23 (50 spots)
- C11 Analyzed through image 29 (47 spots)
Sequential Staining
- imaging new double stained slide, first stain: BX-C-P3 (looks absolutely beautiful, clear high contrast staining).
- kept UV power below 10%. imaged 110K frames.
- should add these to the data collection even if the F06 doesn’t pan out
- added P3-rc toehold probe (more like leg-hold, with 20 bp homology) to solution. Can’t see spots in hybe solution
- all extremely bright spots completely gone in 30 min.
- rehybe with S1 secondary + P3-rc probe to target F06-P1. 1 hr + wash.
- re-add STORM buffer. took some conv and started STORM movie of new test spots. Some substantial background + spots not very bright (as in scarcely detectable on conventional). Wash more, than add more STORM buffer.
- spots still faint. took some conv and STORM movies
- Observation: P3-rc and S1 have substantial complementary structure (10 bp) because we use 10 bp of the common in the S1 binding site. This is not true between P3 and S2 or S4.
- also since all the toehold probes have
- rehybe overnight at 37C heated objective with S1 alone
To try next
- with BXC+ Fo6,
- switch order of probes stained. F06-P3 + BXC-P1
- use just the secondary competetor strand, not the toehold version.
Hybe Prep:
- new hybe dil buffer (1.25x)
- 12 mL dextran sulfate
- 6 mL 20X SSC
- 0.6 mL 10% Tween-20
- 30 mL formamide
- 1.4 ddH2O
Mentoring
- helping Bogdan got sorted with buffers
Thusday 07/03/14
9:50 am – 7:30 pm
coding
- found and fixed bug in
ClusterStats.m
Sequential hybes
- blead-back in of probe signal: probably a sign of not sufficiently sealed chamber. Try air drying sample and then sealing really tight.
- hybe2 stain initially very bright
- hybe2 not very strong at all in previously imaged loci — 405 damage to existing probes?
- non-imaged loci give much better stains
- it does seem the conventional images are still substantially dimmer than they started out. Maybe we should turn off the heater after the hybe
- rehybing probe 2 to confirm this difference.
- need to make more STORM buffer
New stains
- repeat BXC + F06
STORM analysis
- start CC analysis of C09 (eve locus). Looks beautiful.
Wednesday 07/02/14
10:00 am – 11:35 pm
Goals
- Remove panel (c), add multiple examples of 405-cy5 BX-C spots for oligo-secondaries for previous panel (e)
- Fix feducialDriftCorrection — github still preserves the lowercase name.
- maybe best fix is to correct spelling (FiducialDriftCorrection)
- write to Hazen Steve and Shu about fixed adaptive optics demo
STORM imaging sequential staining
- finished staining L3C02, L3E05, L3D08toD12 and sequential stain prep BXC-P3 + F06-P1
- imaging BXC-P3 part
- some issues with stage. Had to use a screw to jammed into the stage from below to defeat the spring loaded catch that holds the stage insert in place (and prevents the stage z-piezo from moving up and down to correct z-drift).
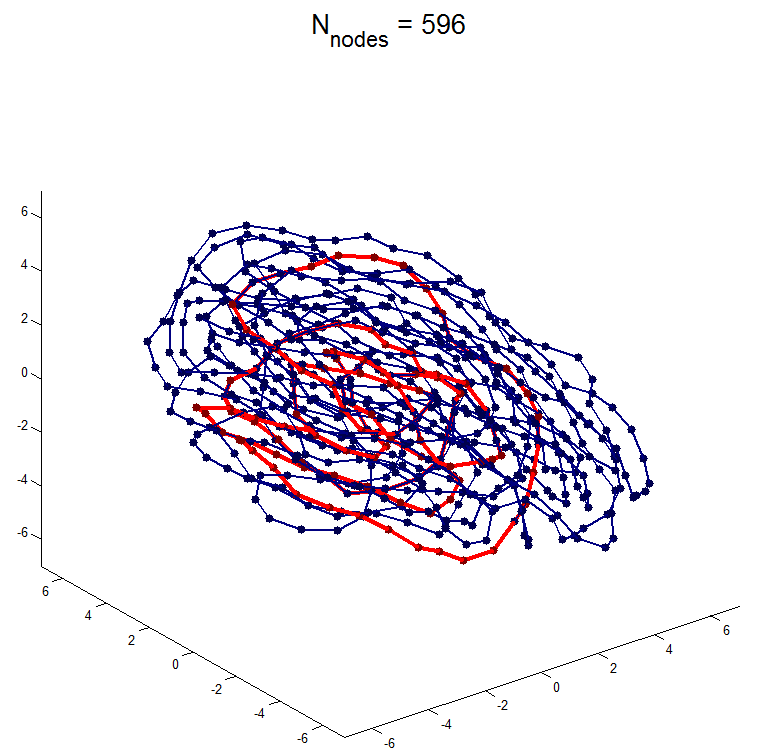
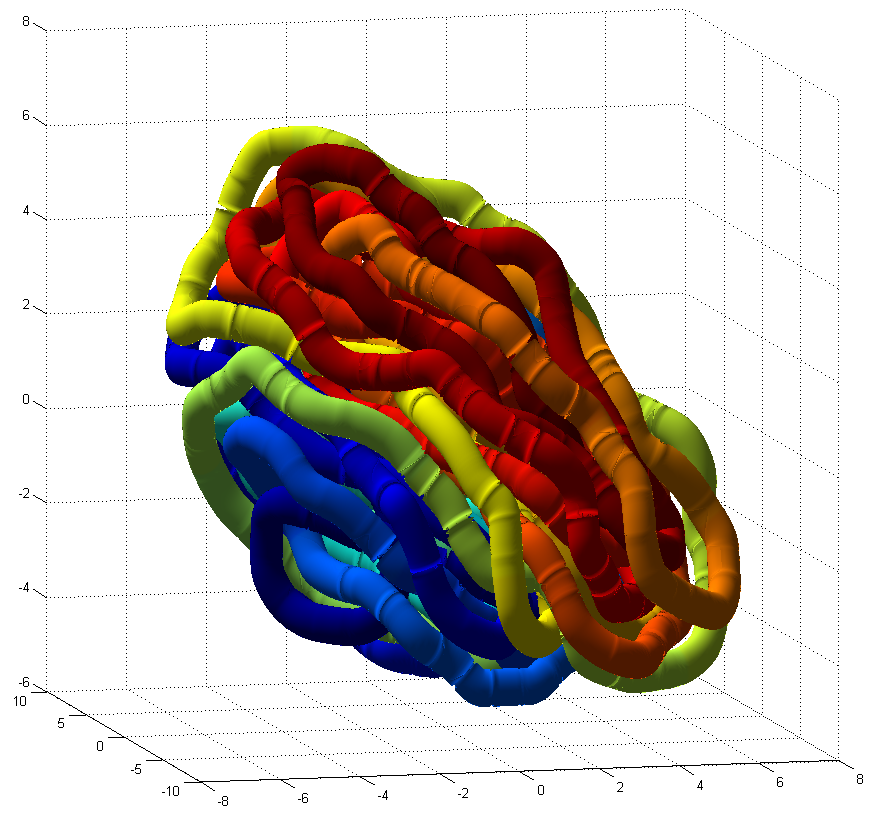
simulation results with adaptiveLangevin
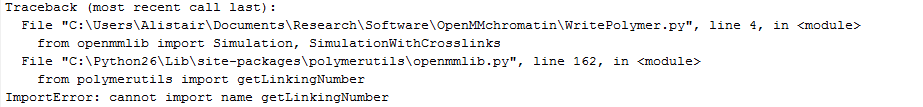
Python path errors
- tried moving code out of site-packages directory. Get the following error:
This makes no sense to me, since it clearly found the polymerutils folder, since it found openmmlib.py which sits inside the polymerutils folder (which is in site-packages, so it certainly should find it). But polymerutils.py and getLinkingNumber is defined inside openmmlib.py
Ah I see the problem: Python doesn’t distinguish the difference between folders and .py files and it is getting confused that there is a polymerutils.py file in a folder called polymerutils\. I suspect it is looking for getLinkingNumber.py inside ...\site-packages\polymerutils\ rather than inside ...\site-packages\polymerutils\polymerutils.py.
Fixed this finally. Code now properly running from external folders.