November 2025 M T W T F S S « Aug 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 Categories
- AP patterning (13)
- Blog (1)
- Chromatin (88)
- Conference Notes (72)
- Fly Work (54)
- General STORM (25)
- Genomics (134)
- Journal Club (22)
- Lab Meeting (66)
- Microscopy (79)
- Notes (1)
- probe and plasmid building (58)
- Project Meeting (3)
- Protocols (13)
- Research Planning (74)
- Seminars (21)
- Shadow Enhancers (59)
- snail patterning (40)
- Software Development (5)
- Summaries (1,412)
- Teaching (9)
- Transcription Modeling (40)
- Uncategorized (10)
- Web development (19)
Links
Tags
analysis cell culture cell labeling chromatin cloning coding communication confocal data analysis embryo collection embryo labeling figures fly work genomics hb image analysis image processing images in situs Library2 literature making antibodies matlab-storm meetings modeling MP12 mRNA counting Ph planning presentation probe making project 2 project2 result results sectioning section staining shadow enhancers sna snail staining STORM STORM analysis troubleshooting writing-
GitHub Projects
Tuesday 07/01/14
7:30 am – 12:25 am
Goals
- finish probe making
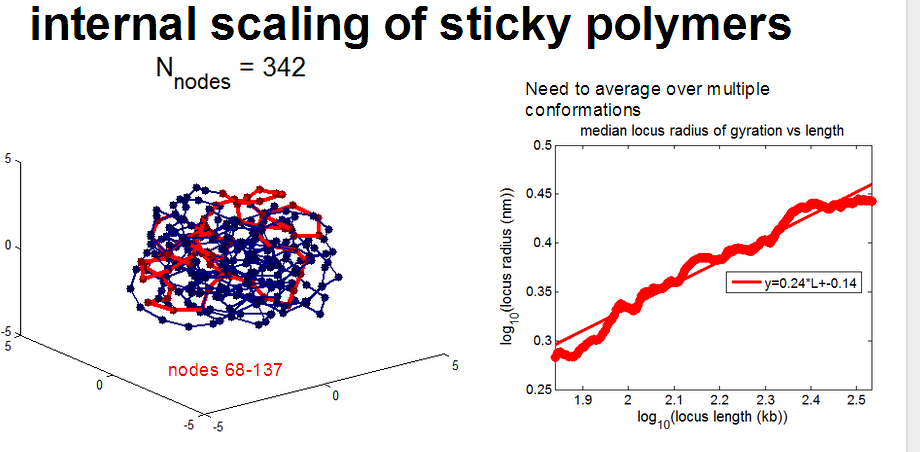
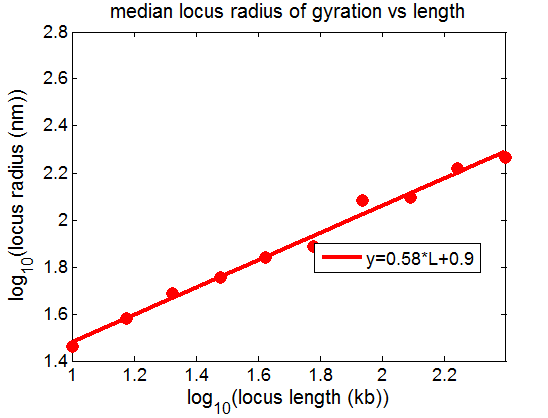
- compute scaling of sub-paths through condensed sticky globules
- revise fig 2 for oligo secondaries
- write caption for fig 2 for olgio secondaries
- write new discussion section corresponding to fig 2 for oligo secondaries.
Oligo secondaries paper
- working on figure 2
- wrote draft caption for fig 2
- working on revising main text.
- working on supp fig 14
- working on XZ’s revisions round 2
Probe making
- label probes,
- test stain new cells with D08-D12-p3
Meetings
- send email to LM
- meeting with Anton, discuss preventing polymer explosions — see notes
- send Matlab Steve viewing code to Hao
Cell staining
- new stains,
- C02-P1-405 + S1-A647,
- E05-P1-405 + S1-A647,
- D08toD12-P3-405 + S3-A647 1.5 uL each probe
- L2F03-05 P3-405 + S3-A647 + L2F06-P1-405 (40 uL) 1.5 uL each probe
Coding notes
- running
MirnyStickyTemplateV6with (global) energyMinimization, variableLangevin integrator, and SmoothSquareWellTailedForce for specific attractions.
a = SimulationWithBonds(timestep=20, thermostat=0.05)
a.setup(platform=platform, verbose=True, GPU=GPU,integrator="variableLangevin",errorTol=.0001)
SimulationWithBondsinherits from Simulation and I’ve just added my own potentials here rather than modify openmmlib.py.
Does it matter what I put for timestep if I’m using “variableLangevin” integrator? Have I specified the error tolerance for the variable integrator correctly?
Posted in Summaries
Comments Off on Tuesday 07/01/14
openMM simulation notes
Recommendations from Anton for avoiding exploding polymers
Initial recommendations
- integrator choice:
- variableLangevin integrator recommended
- takes an error tolerance instead of a timestep variable (e.g. 10^-4)
- thermostat = high values high friction
- mass_scale = argument of init of simulation (see openmmlib.py)
- attractionRadius (be careful about interaction over other polymers).
- wiggle_dist = higher more oscillation, less chain crossing.
- localEnergyMinimization(maxIterations=1000,tolerance=1)
- local energy minimization with attractive potentials might lead to super-position.
- Try global energy minimization.
conlen = 1 nm * length_scale (=1).
potentials
- smooth square well force
- polynomial force
- sticky particles should be extra hard.
- every particle has a charge (sticky yes no) repulsion strength yes no.
- extra repulsive force
- try hard-core forces (very large potential energy at overlap).
dynamic adjustments (changing parameters during simulation)
- things of interest to change: friction
- crude way, use output as input
- system block
- try energyMinimization in place of localEnergyMinimization
- avoid any collapsed pairs (they just get worse). — this is time to quit
Monday 06/30/14
10:00 am – 5:45 pm, 10:00 pm – 1:15 am
Meetings
- group meeting (see notes)
- journal club (see notes)
- discuss probes with George
- setting up meeting with AG to discuss code
- discuss with XZ Brian’s paper and revisions
- project 2 team meeting discuss data analysis
Coding
- integrate new cluster analysis code into project 2 core analysis scripts
Probe making
- set up RT reactions
- run diagnostic gel. — clearly have RNase problems substantial smears and reduced incorporation.
Posted in Summaries
Comments Off on Monday 06/30/14
Journal Club: B-Actin mRNA in neurons (Singer)
Science
Basic observation
- chemical stimulation of neuron, see more B-actin mRNA in axon (2x)
- also see more B-actin mRNA following gentle protease treatment — propose mRNA was masked
- do transcription controls and trafficking controls
- block neural stimulation pathway, also don’t see more B-actin
- kinetics of response (detected in less than 10 minutes). within 20-40 min has returned to masked state. (half-life tau = 7 min)
- is this long enough to make bigger spines? Well certainly if you have multiple stimulations
- is more actin actually made? Use dendra-2-B-actin photo-conversion to detect newly synthesized B=actin.
- rRNA detection also increases following protease treatment
- masked B-actin (revealed by protease) associate more the rRNA granuales (revealed by protease).
- bright ribosomal structures have reduced mobility, become more mobile following cLTP
- live imaging of B-actin mRNAs observe faster diffusion in stimulated cells.
Posted in Journal Club
Comments Off on Journal Club: B-Actin mRNA in neurons (Singer)
Protected: lab meeting 06/30/14
Posted in Lab Meeting
Comments Off on Protected: lab meeting 06/30/14
Sunday 06/29/14
10:30 am – 3:00 pm, 6:00 pm – 7:00 pm, 8:30 pm – 11:30 pm
STORM
- finish imaging C11
Cell prep
- passage newly plated cells to 75cm2 flask
- fix cells, prep cells for staining
Probe making
- run RT PCR for D08 – D12 + neg cntrl
- run PCR cleanup
- run IVT reaction
Chromatin Analysis
- Analyzed B10 up through image 23
- Analyzed B11 except last 3 images (7, 8, 9)
- Analyzed B12 through image 4
- Analyzed F11toF01 through image 26 (50+ spots)
- Analyzing F01 nearly same size as F11 to F01 ( through image 31, 43 spts)
- Analyzing E12 much smaller than F01 (despite 2x more DNA, 10kb vs 5 kb),
Posted in Summaries
Comments Off on Sunday 06/29/14
Saturday 06/28/14
10:45 am – 5:30 pm, 9:00 pm – 11:50 pm
Small tasks
- restart data copying after computer freeze last night (GPU computation abort failed)
- restart sticky polymer simulations
STORM imaging
- imaging sample C10 (blue 33 kb vg locus).
- Conv images nice and bright
- images look pretty good.
- STORM imaging sample C11 (black 10 kb)
Meeting with Ajaz
- follow up to ANTP PH cluster analysis: BX-C and control region
- Ajaz found new potential polymer modeling collaborator (see email)
- will get manuscript to XZ
Chromatin Data analysis
- Analyzed D01 data (ChromatinCropper)
- Analyzed D02 data
- what happened to D03 data?!
- Analyzed D04 data
- Analyzed D05 data
- Analyzed D06 data (way too large, not sure what’s up with this locus!)
- Analyzed D07 data
- black trends looking decent
Cell culture
- passaged cells yesterday
- start new cultures from frozen stock today
- density looking pretty good, should be able to plate a batch of these cells tomorrow and passage the rest.
Chromatin experiment planning
- ordered toe-hold displacement probes to try to remove un-bleached, un-imaged secondaries for sequential hybe approach.
Friday 06/27/14
10:45 am – 11:00 pm
Prep for meeting with XZ: model summaries
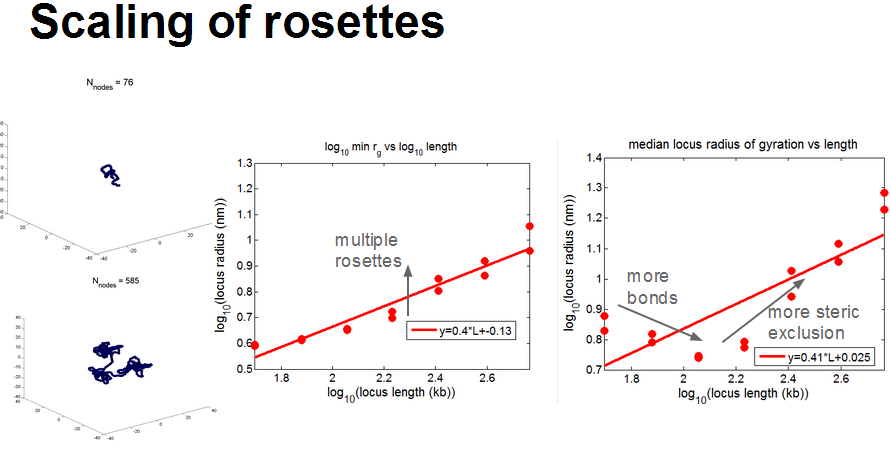
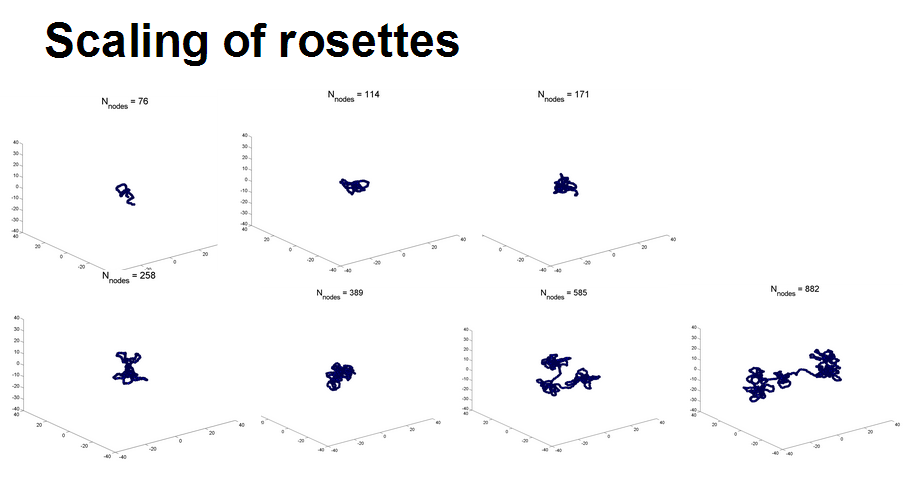
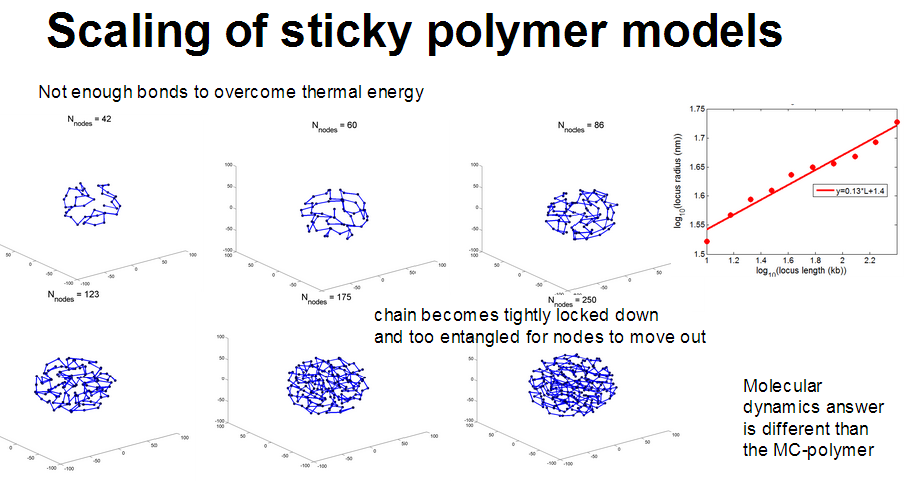
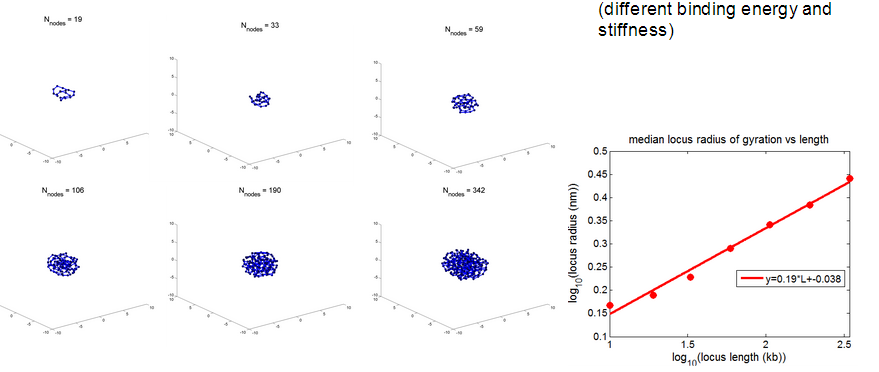
- rosettes actually have very complicated scaling
- generally sticky nodes seems to scale better.
Conclusions from meeting with XZ
- First priority: get at least a first look at the internal scaling of black domains
- Second priority: talk to Hazen and Steven about demoing commercial adaptive optics (improve 750 imaging deep in cells?)
- start discussion with Mirny group
Oligo secondaries
- working on figure
- find some regions to do cross-sections?
STORM
- finish staining C09, C10, C11 (hot washes)
- imaging C09
Posted in Summaries
Comments Off on Friday 06/27/14
Thursday 06/26/14
9:45 am – 1:30 am
Chromatin Modeling continued
Checking my simulation results from the polymer models I was running last night.
Simulation Results
BBv1 no attractive potential does give expected scaling:
Adding an attractive potential does increase clustering. Has more effect on large polymers.
Notes from meeting with Max
- Combine repulsive and attractive forces into a single potential.
- use modified square well
- Need to learn better use of python classes and namespaces. See Python class doc: https://docs.python.org/2/tutorial/classes.html
More coding
- Fixed up openmm-polymer code
- implemented extension script to import and build off the openmmlib (I called my function lib
openmmlibExt). - with some difficulty (stupidity), finally implemented node specific interactions
- simulating free polymers,
- simulating uniformly sticky polymers
- simulating periodically stick polymers
Primer ordering
- Lib4 primers still had the same bug as the Lib3 primers. Now when back and editted old scripts to fix this bug (will be logged in GitScripts).
- bug was in the export list of T7 rev primers, the sequences of the Fwd primers was used and not the rev primers.
- wrote new functions to index 96 well plates and to convert fasta files to csv tables for ordering primers.
- validated primers in quick test.
- remembered (belatedly) to add T7 to project 2 L5 primers and strip off extra Gs
- sent primer orders to Alec.
Staining
- new cell stains: C09 C10 and C11 (tiny blue and black regions)