9:00 am – 7:10 pm, 9:00 pm – 10:00 pm,
Goals
- finish lab meeting presentation for tomorrow
- finish CONTE center presentation for tomorrow
- finish update slides for discussion tomorrow
Analysis
Morning tasks
- check STORM imaging
- movies still going well
- issues with beads in this sample, 561 is suddenly backgroundy but not a lot of beads in the focal plane.
- Auto-analyze on arrival
- didn’t work. Changed defaults in RunDotFinder, seems to be working now.
- communication with CW, SN and DD about oligos and Biosearch/Stellaris
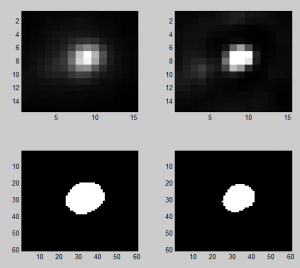
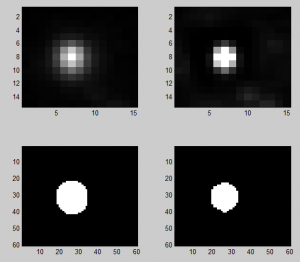
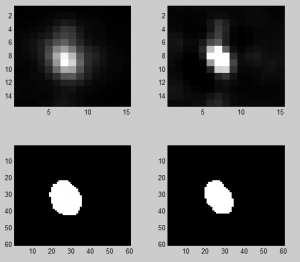
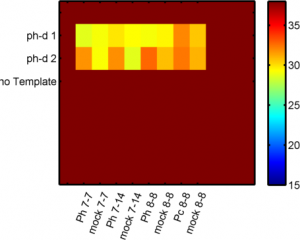
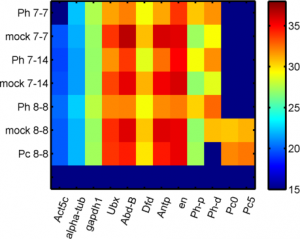
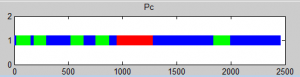
Analyzing Ph data
- cleaning up data
- converting BB’s molecule lists back to standard molecule lists
- need to convert xc and yc back to pixels
- need to convert molecule lists back to structures of cell arrays rather than large structures
- need to compute imborders
- BB flists keep crashing my 3D plotter — haven’t converted everything very well yet.
File Organization
- Old TSTORM folder on my embedded DATA drive (D) is taking up too much space — consoldiating this on to the TSTORM data drive
- need to remember to delete after it finishes copying the ~600,000 tiny files over.
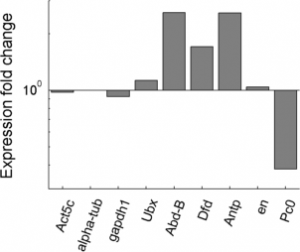
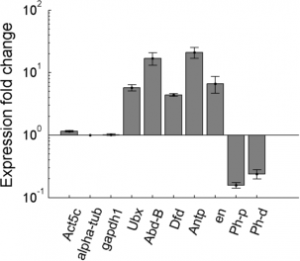
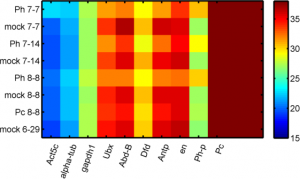
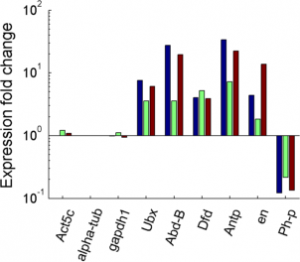
Imaging Ph in Ph-KD
- some difference in overall brightness — hard to quantify nicely since there is clearly background autofluorescence that needs to be accounted for.
RNAi NextSeq data
- data arrived today.
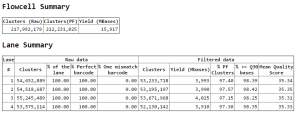
- Running Bowtie
- error 1: mapped to two-stranded genome, all reads map multiple times
- error 2: mapped to positive strand genome, 80% unique reads, 20% multiple alignment. But names need to match GTF file
- need to match expected folder structure — added these notes to my pipeline script
- would be nice to add cufflinks into this pipeline.
- need Bowtie Genome fasta to match reference names in genome GTF file given to cufflinks
- SAM file needs to be sorted — converting to BAM and sorting using sam tools
- installed same tools for windows from here
Analyzing PhKD data
- serious lab issue on Monet. Going for the reboot. Hoping the Windows 10 install plan doesn’t kill it again.