9:30 am – 8:15 pm, 9:30 pm – 11:30 pm
STORM
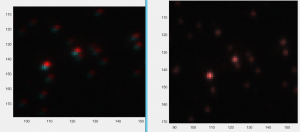
- continued imaging ANT-C G6 in WT cells (imaging finished at 6:17 pm)
Staining
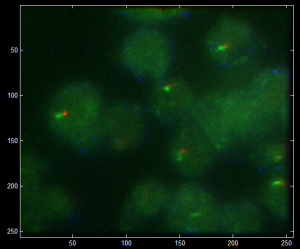
- rinsed new antibody stains (CycA and Ph)
- CycA (after formamide) now only stains the cytoplasmic fraction
- 488 is extremely bright
- still distinguishable by confocal (not so obvious by eye-pieces).
- This Ph still is not nuclear in these Kc cells
Ph troubleshooting
- added 60 uL blocking buffer to dried out / empty tube of old Ph antibody. Maybe there’s still some left. Used 20 uL of the resuspended anti-serum to stain WT cells.
- incubated 1 hour at room temp and then overnight at 4C
STORM
- CycA stain is essentially undiminished from this morning. Still very bright. C03 stained clearly as well
- BXC stained well but the replacement F06 I used did not stain well
- reverting to the previous sample (here BXC is a little below what I desire and in 750, though F06 is bright).
- tomorrow I should try staining BXC-P1-647 + F06-P3-750.
Fly husbandry
- flipped stocks.
Presentations
- working on lab-meeting presentation for Friday
Communications
- send Hi-C summary to XZ
- reply to CW about cell cycle
- reply to NF about paper