10:30a – 11:45p
Data analysis
- Launch STORM analysis for bead data
- crazy transpose now between conventional images and STORM images in F02F03 data. May affect all recent data?
To do:
- Finish fellowship progress report
- 2 color imaging of other en-locus boundary regions
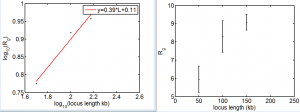
- draft fig 3 for project 2 to show XZ + meeting
- analysis of 2 color
- fix new cells for new multi-color stains and continuing analysis of more yellow regions
STORM imaging
- multi-color imaging F04F02
- cell 0001: 1 uL pure COT + 2.5 uL MEA. Not too bright.
- cell 0002-0003: brand new glox. 10 uL DMSO dissolved COT. 2.5 uL MEA
- beads clearly bleaching fast.
- domains look spread out / way too big in 647.
- single-color imaging O/N L3B04
Cell culture
- cells a bit over-grown, not passaged for nearly a week.
- passage cells
- plate new cells for fixation
- original plating density a bit to high. removed excess cells and added new media.
Modeling
- OpenMM tutorial with Bogdan
- implemented spherical confinement potential. See tuck
c:\polymer\for code.
New stains
- F06p1-F05p3, F03p1-F04-p3, no secondary.
- B05-p1 + .5ul A647
- plate 40 mm cells