9:45am – 4:45pm,
Goals
- Set up RT reactions Lib3 samples B10-B12, C09-C12, D01-D08
- Wash newly stained samples (B08, B09, E11-F01, + PH immuno-FISH)
- set up STORM imaging of PH immuno-FISH
Lab Meeting
General
- letterhead and signature for letter for NF
PH Project
- test GA-postfixed cells. no staining of chromatin loci. higher 561/488 background (probably from GA).
Chromatin project
Probe making
setup RT reactions
- per reaction
- 4 uL of P1 A405.
- 5 uL of RT buffer
- 2 uL dNTP
- 14 uL RNA
- 1 uL RNase inhibitor RNAsin
- .25 uL Maxima
- master mix 16x
- 64 uL P1 A405
- 80 uL RT buffer
- 32 uL dNTP
- 16 uL RNase inhbitor
- 4 uL Maxima
Results
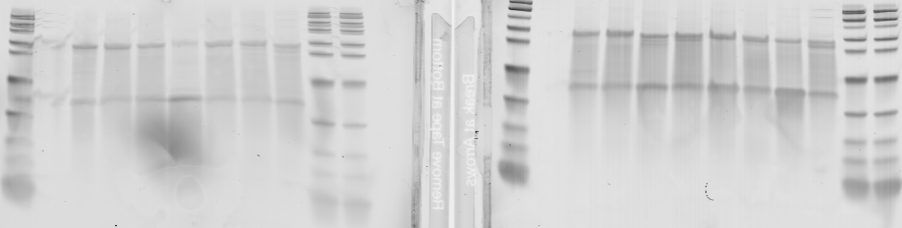
- ran RT reactions (30 uL scale, 400 uL primer, 15 uL RNA). (B10-B12, C09-C12, D01-D08).
- poor incorporation and some smearing I think I’m getting substantial RNase degradation. Maybe should run the T7 reactions shorter and let it sit for less long before running RT.
- probably still decent enough for a staining, we’ll check the concentrations and probably dilute a bit less onto cells.
Left B10-B12, C09-C12, Right D01-D08.
STORM
- STORM imaging of Y B08 region.
