November 2025 M T W T F S S « Aug 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 Categories
- AP patterning (13)
- Blog (1)
- Chromatin (88)
- Conference Notes (72)
- Fly Work (54)
- General STORM (25)
- Genomics (134)
- Journal Club (22)
- Lab Meeting (66)
- Microscopy (79)
- Notes (1)
- probe and plasmid building (58)
- Project Meeting (3)
- Protocols (13)
- Research Planning (74)
- Seminars (21)
- Shadow Enhancers (59)
- snail patterning (40)
- Software Development (5)
- Summaries (1,412)
- Teaching (9)
- Transcription Modeling (40)
- Uncategorized (10)
- Web development (19)
Links
Tags
analysis cell culture cell labeling chromatin cloning coding communication confocal data analysis embryo collection embryo labeling figures fly work genomics hb image analysis image processing images in situs Library2 literature making antibodies matlab-storm meetings modeling MP12 mRNA counting Ph planning presentation probe making project 2 project2 result results sectioning section staining shadow enhancers sna snail staining STORM STORM analysis troubleshooting writing-
GitHub Projects
Sunday 03/30/14
10:40a – 12:15a
Some basic organization
matlab-functions collaboration
- discussed plans
- make headers match more strictly
- move all user prompts to pop-up boxes not command line prompts
Project organization folder structure
- change all scripts folders to short-cuts or symbolic links to the GitScripts folder.
Backups
- pushed GitScripts
Chromatin Project stuff
Cell staining
- rinse out primaries from E01-E03 stains
- move to 4C, will image tomorrow night.
Chromatin Analysis
Analyzing F03-F05 data from 01/17/14
- overall data looks pretty good
- some alignment / drift / conventional-storm registration issues at cell 8 or 9.
Analyzing F04-F03 data from 01/15/14
- some cells look pretty good
- 750 counts drop pretty rapidly after the calibration movie.
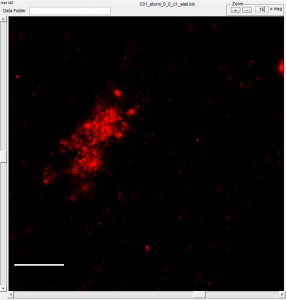
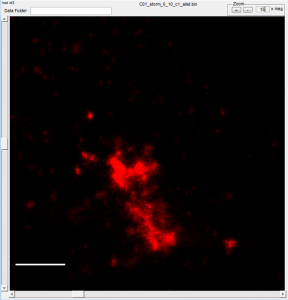
- some cells have clear alignment mismatch / drift issues. See conv vs STORM:
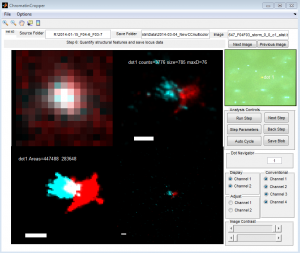
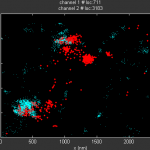
Data so far
- exploring new plotting options
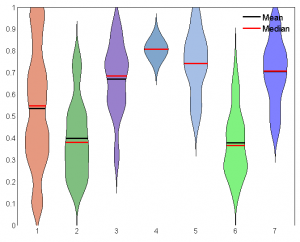
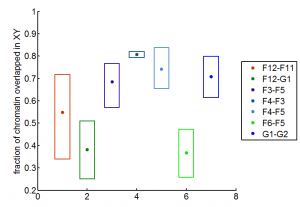
- violin plots for matlab — somewhat useful.
- also interesting, distributionPlot. Added to my FromFileExchange functions.
- some comparisons between blue-blue, blue-yellow, and black-yellow
Ideas for further analysis
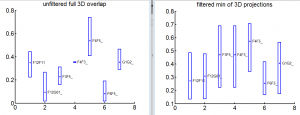
- screen through good 3D data, provide 3D overlap analysis.
- Surprising trends from 3D analysis
Ph Project, to do:
- Make supplement image of antibody controls
- write caption for supplement image of antibody controls
- remake supplement figure on counts vs. diameter to capture new standards (now it’s own proper figure script).
- write caption for supplement image of molecule counts (instead of diameter) – complete
Posted in Summaries
Comments Off on Sunday 03/30/14
Saturday 03/29/14
10:15a – 12:15a
Multi-color data analysis
- Analyzing chromatic bead data from 2014-02-27 and 02-28
- Analyzing drift bead data from 2014-02-27 / 28
- should write a little script to auto launch this stuff?
- 2014-02-28 F06F05 dataset, Image 25 and on, Conventional no longer aligns with STORM image :(. Still have 50+ excellent images.
- 2014-02-27 F04F05 dataset — 647 F5 very poor switching, few localizations. Overlap still looks reasonable.
- worth analyzing but also worth repeating.
Probe Making
- Set up new RNase free area in upstairs lab with new dedicated pipettes.
RT reactions
- E01 – E05 with P1 (tubes 1-5)
- E03, E04, E05 also with P3 (tubes 6-8)
- E06 – E08 failed PCR, try again in a later batch.
- Out of P3-405!
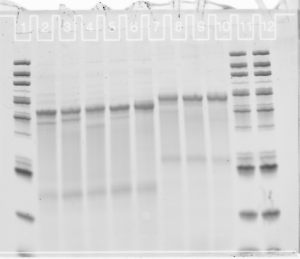
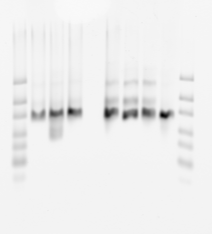
denaturing gel
- loading order E01-E05 P1, then E03-E05 P3.
Final probe concentrations
Ph Project, to do:
- Make supplement image of antibody controls
- write caption for supplement image of antibody controls
- write caption for supplement image of molecule counts (instead of diameter)
Friday 03/28/14
9:45a – 11:50p
Goals
- Record bead fields.
- check isoforms differences and consequences for probes selected (do our probes still hit the most popular isoform of imr90s?)
Cell Staining
- washout stains for new multi-color samples. Need to wait until oven is free.
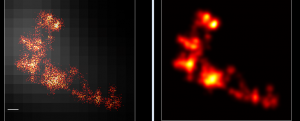
Imaging samples
- F03-p3 2 uL, F02-p1 7 uL — F02-P1-A647 actually works (first time!) should image this. F03-P3-cy7 is detectable but very dim. I have gotten much brighter A750 stains before. Background is also high.
- F05 P3 1.5 uL, F06-P1 6 uL — F05 probe synthesis failed. F06-P1 looks strong and clear at thic concentration.
- F06 P3 2.5 uL, F07-P1 6 uL — F06-P3-cy7 failed ? High background. F07-P1-A647 looks fine. Not clear why F06 works with P1 and not with P3.
New Stains
- F3-6, F1-7 6 uL, 6 uL
- G6-6, G8-7
- G8-6, G6-7
- F7-6, F6-7
- F6-6, F7-7
- G2-6, G3-7 1.5 uL, 1.5 uL (~1100 ng/uL)
New Probes
- E01-E08
- chr3R:19929199-20187132__YELLOW_250kb_silent_flanks
- chrX:1907754-2083414__YELLOW_175kb_silent_flanks
- chr3R:19929199-20029199__YELLOW_250pt1_100kb
- chrX:1907754-1977754__YELLOW_175pt1_70kb
- chrX:1977754-2047754__YELLOW_175pt2_70kb
- chr2R:19726615-19787585__YELLOW_D12a
- chr2R:19809874-19888410__YELLOW_D12b
- chr2R:19906491-19976552__YELLOW_D12c
- Neg control, E01fwd, E07rev
- ran PCRs
- started T7 reactions
Not sure why E06 and E07 failed, and E08 looks weird. Maybe press on with just E01-E05.
Research Organization
- moved ChromatinCropper into matlab-functions
- moved commonly used mature functions out of Beta into appropriate matlab-functions folder (e.g. normhistall is now in Plotting).
- moved storm-analysis and storm-control into Software.
- moved Scratch into Software
- moved GeneralSTORM scope images onto Data and archived code into Archive2014 on D drive
Thursday 04/27/14
10:15a – 12:30a
Goals
- Data processing
- copy C05 and C06 data to hard-drives
- Launch analysis of C03-C06 data
- plate, fix, prep and hybe new cells
- Probe making
- RT reactions
- test gel
- probe purfication
- cell staining
- wash out probes
- image multi-color cells on STORM2
Chromatin Project
Probe making.
- finished making 16 new probes (2 failed, no template).
- gel looks pretty uniform for concentration
- F03 has weird series of bands — probably not good. Should remake. F05 failed completely (no template, I guess I knew this. Good negative control).
- attempted 500 pmol scale synthesis. Incorporation looks less good, should stay at 400 pmol per 10 uL RNA.
Cell staining
- fixed and prepped new cells for staining, including 1hr+ RNase A treatment at 37C.
- F03-p3 2 uL, F02-p1 7 uL
- F05 P3 1.5 uL, F06-P1 6 uL
- F06 P3 2.5 uL, F07-P1 6 uL
- Damn it I used the failed and questionable probes in the staining. Need to check these things!
- Why do I get better yield on the failed probes?
Imaging
C07 on STORM2
- 3 uL COT, 5 uL BME,
- looks reasonable
- Need to remember to take beadfields in the morning! Otherwise there will be lots of beads to manually parse out of ChromatinCropper.
C08 on STORM1
- 2 uL COT, 5 uL BME
- spots identifiable in all cells, but weak / small / low contrast.
- realigned back-optics (way off center)
- tweaked dual view to separate channels Cy5Cy3 3D. Alignment is off by 10s of pixels between channels.
- Need to remember to take beadfields in the morning! Otherwise there will be lots of beads to manually parse out of ChromatinCropper.
Coding / Research Organization
- established GitScripts repo on github, pushed scripts. Now these and their revision histories backed up somewhere. Yay!
- pulled latest version of script databases into GitScripts and committed and pushed up to Github (This takes a couple commands, including a move command to get new scripts to there appropriate folders. I bet I can write a matlab wrapper to do this in a single click.)
- added private protocols to GitScripts
Posted in Chromatin
Tagged cell staining, chromatin, images, probe making, STORM
Comments Off on Thursday 04/27/14
Wednesday 03/26/14
9:20a – 10:10p,
remotely, 10:50p – 11:30p
Goals
- passage cells
- washout probe from new multicolor stain
- finish updating notes from yesterday
- STORM run configurations
- data analysis launched
- break down PRISM2 run of L3C04. (complete)
- break down STORM2 run of L3C03 (complete)
- set up copying and data-fitting of L3C03 and L3C04 data (copied, started fitting C03)
- start making new probes from Lib2: F01, F02, F07, G06, G08, G09
Longer goals
- More probe making
- lower priority, remakes: F06, G05,
- Lib3 on deck: embedded yellow regions E03-E10
- start ChromatinCropper of L3C01
- ChromatinCropper of F05-F06 02/28 data (and 02/27 data?)
- analyze buffer data
Chromatin
Imaging
New multicolor stains
- New stains failed. I believe I previously verified the low concentration (1.5 uL probe) of these probes makes this happen. Repeat at higher concentration.
Sample C05 on STORM2
- 3 uL COT, 5 uL BME buffer
- staining looks pretty good, spots look decent.
Sample C06 on STORM4
- 5 uL COT, 5 uL BME.
- dots are clearly fainter
- switching pretty well, background is tolerable but a bit higher than desired for imaging small (<50kb) chromatin regions.
Cell Staining
- Need to stain new cells today if I want to take adventage of the microscope time I have this week.
- Despite just passaging cells, between the two culture flasks I think I easily have enough cells to plate another 12 coverslips. Let’s fix new cells tomorrow.
- New stains: C07, C08, and double stain F06-A647 + F05-A750.
New probes making
- Lib2: F01, F02, F03, F05, F06, F07, G06, G08
- ran PCR. F05 sublibrary failed, rest look good. Press ahead with all
- ran PCR clean up / DNA clean and concentrate 5.
- setup 20 uL T7 reactions to run overnight.
Data Analysis
- all data running last night finished except the 02/28 double color data (which I was excited to analyze). This is still chugging along at a conservative 25% CPU on Cajal.
- canceled and relaunched analysis with 8 parallel processes per color. Along with the C03 analysis this has Cajal up to 100% CPU engagement overnight again.
Ph project
- email Nicole clarification and explanation of graphs.
- further email discussion and clarifications
- To do:
- normalize all distribution plots. Report both total Ph localizations and total clusters.
- consider truncating distributions at 700 rather than clustering into last bin. It’s important this doesn’t look like a second peak.
- add full length distributions (out to 1.5 um) into supplement.
Tuesday 03/25/14
10:00a – 1:00a
Oligo Secondaries
- Fixes for secondary figure
- use the dots only rendering method
- make dots only rendering method a standard plugin for matlab-storm (I rather like it)
- wrote function
StormHist.madded to QuickPlots of matlab-functions instead, not sure how general this will be. - downsizing the resolution of the STORM image of the simulation to simulate conventional image. This seems to work well, however the image ends up poorly centered (center is shifted to upper left as zoom get less). Can’t figure out why this is.
- As nice as this with list2img and STORMcell2img, its actually a pretty simple function and maybe would be better to write from scratch as a stand alone call of hist3.
- Sent current draft version back to Brian.
- Should be good enough for writing, I don’t propose changing the storyline of the figure at all, just tweaking the actual images a bit.
- I like the decompacted structure in this version better.
Ph projects
- comments back from Kingston.
- saved to disk. haven’t read yet.
- Ajaz revising manuscript tactics with Nicole, will write later.
Chromatin
Staining
- Forgot to do melt step on new multicolor stains last night! executed this morning at 9:30p
Data Processing
- Launch analysis for L3C01 dataset
- really should re-institute gain into bead images, bead image quality is marginal without it.
- 2014-02-27_F04-6_F05-7
- data is all unanalyzed. Processing now on Monet.
- switching looks like crap.
- 647 in particular everything is high-count low contrast and switching is horrible.
- this is very likely the consequence of the mistaken preparation of 1M MEA in H2O instead of in acid
- 750 data also looks poor, though not as bad relative to its norm as 647.
Data Organizing
- Finished consolidating all multi-color data from ProBox1-7 onto ProBox8
- To do: move multi-region single color data from ProBox8 onto ProBox7. This will help reduce confusion and make space for the multi-color data from RAID2 to go onto ProBox8.
Imaging
Sample C02 imaged on prism2 (started last night)
- O/N run on PRISM2 severe focus lock error at 8:45a (I wonder how good the images are from before then, this focus lock seems pretty happy to record off-set zero at a variety of different focal planes).
- Finish imaging on PRISM2 at 11:00a.
- several focus lock errors on PRISM2, need to check data for quality — might need to repeat this imaging session.
Sample C04 on prism2
- Check Prism2 laser powers
- 657 has 70 mW at back appeture (not bad, though this line is rather off peak spectrum)
- 405 about 200 uW, set to ND filter position 3. Previous user has been blasting with 100% UV. This leaves my dyes permenantly on even at <1% AOTF laser power.
- attempted to add auto gain. Not working. (note
^does not exponentiate in python. Need to use**). - hopefully these beads are still usable (with a dynamic range of only 16!).
sample C03 on STORM2
- laser powers 65 mW at backport with GUI set to 300 mW 647
- attempted to branch storm2 and pull in DaxWriter-master from alistair storm-control. Resulting Hal configuration is non-functional.
- switched back to old NewDaxWriter branch. This seems to still work fine.
A quick look at the C01 data
Project 2
- see protected notes
Computer upgrades
- Call CDW about getting windows 2012 quote with HHMI billing address — no reply, left message
- academic license from NewEgg seems to quote in at considerably higher (600+ instead of 230). Not entirely clear it’s the right license either.
Literature
- Finish reading Johnston and Desplan Science paper.
Protected: project 2 update and planning
Posted in Genomics
Comments Off on Protected: project 2 update and planning
Monday 03/24/14
10:00a – 11:15p
Chromatin Project
Data summaries
- making slides to summarize recent analysis of multi-color data
- see slides for 3/28/14 XZ meeting
Data analysis
F03-F04 data
- chromewarps obviously don’t match data, clear multi-pixel shift up and to the right on all spots
- Split up the many steps of CalcChromeWarp into separate functions (finally) so that they can be used independently.
- This still needs some improvement given current variable names / data storage formats, but its a step in the right direction.
- modifying ChromatinCropper to change warp files via the Menus and ‘remove last blob’ via menu commands.
STORM
PRISM2 coding
- branched off from prism2. Pulled daxwriter-master (pushed up from STORM4) down into new branch.
- New version of Dave won’t work with Steve until Steve is brought up to speed with new TCP protocol communication method
- updated steve-settings using the prism2 branch defaults.
imaging observations
- imaging C02 sample on PRISM2
- using Cy5Cy3 3D dualview insert.
- laser beam needs some alignment, not too bad, I’ll just use it as it is today
- system loses focus — offset will got to -1300, and stay there, despite lock target set at 0.
- molecules quite dense (high background?) and staying on for multiple frames (higher laser power would really be nice).
- IR focus lock sum signal varies between cells. This system has high sum signal values (0 – ~1600? instead of 0-255).
Genome patterns
- enhancers don’t cross boundaries – example brk enahancers inside likely contiguous yellow domain flanked by likely blue / black regions.