November 2025 M T W T F S S « Aug 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 Categories
- AP patterning (13)
- Blog (1)
- Chromatin (88)
- Conference Notes (72)
- Fly Work (54)
- General STORM (25)
- Genomics (134)
- Journal Club (22)
- Lab Meeting (66)
- Microscopy (79)
- Notes (1)
- probe and plasmid building (58)
- Project Meeting (3)
- Protocols (13)
- Research Planning (74)
- Seminars (21)
- Shadow Enhancers (59)
- snail patterning (40)
- Software Development (5)
- Summaries (1,412)
- Teaching (9)
- Transcription Modeling (40)
- Uncategorized (10)
- Web development (19)
Links
Tags
analysis cell culture cell labeling chromatin cloning coding communication confocal data analysis embryo collection embryo labeling figures fly work genomics hb image analysis image processing images in situs Library2 literature making antibodies matlab-storm meetings modeling MP12 mRNA counting Ph planning presentation probe making project 2 project2 result results sectioning section staining shadow enhancers sna snail staining STORM STORM analysis troubleshooting writing-
GitHub Projects
Protected: project 2 update 03/24/14
Posted in Genomics
Comments Off on Protected: project 2 update 03/24/14
buffer tests 04/23/14
- 1% BME
- 10 mM MEA
- 0.5% BME + COT
- 2% BME + COT
- 10 mM MEA + COT
- 50 mM MEA +COT
- 50 mM MEA + SSC + COT
Posted in General STORM
Comments Off on buffer tests 04/23/14
git remove binary files
See current file size. First need to git gc
Alistair@MONET ~/Documents/Research/Projects/Chromatin/Code (master)
$ git gc
Counting objects: 713, done.
Delta compression using up to 8 threads.
Compressing objects: 100% (570/570), done.
Writing objects: 100% (713/713), done.
Total 713 (delta 257), reused 459 (delta 142)
Now we can count objects. size-pack is the size of our database in Kb (70+ Mb here!)
Alistair@MONET ~/Documents/Research/Projects/Chromatin/Code (master)
$ git count-objects -v
count: 7
size: 22
in-pack: 713
packs: 1
size-pack: 70827
prune-packable: 0
garbage: 0
Here we get rid of the offending file. For more details on all of this, including a good guide on how to find the offending big file(s), see this part of the Git Pro Book.
Alistair@MONET ~/Documents/Research/Projects/Chromatin/Code (master)
$ git filter-branch --force --index-filter \ 'git rm --cached --ignore-unmatch
mRNAdata.dax' HEAD
Rewrite 4c0556f7efab2e9e23fa09dfd247edabfc6ef301 (1/46)rm 'mRNAdata.dax'
Rewrite 7ec96fb9c5ba3d23d4a55022f2a094635835901e (2/46)rm 'mRNAdata.dax'
Rewrite bba4dfa4d06cf51b06a8ba937a84ef4700f44094 (3/46)rm 'mRNAdata.dax'
Rewrite d6a21db5bd63f1c1a23f5633278e84db8763348e (4/46)rm 'mRNAdata.dax'
Rewrite c25106ae199f3a84024a9609434ca9b6818cf632 (5/46)rm 'mRNAdata.dax'
Rewrite 3214522b942f96b579d15bf1b95c2e8daeed0d36 (6/46)rm 'mRNAdata.dax'
Rewrite 94f2d457c83b45ef14b01b479c59e8a94c262d2f (7/46)rm 'mRNAdata.dax'
Rewrite 43d60c2c03fa9c1125148e0b8ca22b8c2d7d622d (8/46)rm 'mRNAdata.dax'
Rewrite a0c181e158a125549b2ac80a48058bb7f075a9ec (9/46)rm 'mRNAdata.dax'
Rewrite d63f8f7ff072f836685eaadbb71128be79ea4186 (10/46)rm 'mRNAdata.dax'
Rewrite 9f64ccff4b1de60bea49b3e74020305768568df2 (11/46)rm 'mRNAdata.dax'
Rewrite 83c96e3fbe80cc3ca17b2b5c847488576081aeb6 (46/46)
Ref 'refs/heads/master' was rewritten
Now some cleanup. We remove the refs and logs
Alistair@MONET ~/Documents/Research/Projects/Chromatin/Code (master)
$ rm -Rf .git/refs/original
Alistair@MONET ~/Documents/Research/Projects/Chromatin/Code (master)
$ rm -Rf .git/logs/
And we want to remove the rewrite backup so that everything is back to normal. Otherwise if we need to do this again we will get errors objecting to our editing the rewrite folder. I haven’t found as good references to support this move so maybe do with caution.
Alistair@MONET ~/Documents/Research/Projects/Chromatin/Code (master)
$ rm -rf .git-rewrite/
Now let’s see how we’re doing on space
Alistair@MONET ~/Documents/Research/Projects/Chromatin/Code (master)
$ git gc
Counting objects: 712, done.
Delta compression using up to 8 threads.
Compressing objects: 100% (463/463), done.
Writing objects: 100% (712/712), done.
Total 712 (delta 257), reused 655 (delta 248)
Alistair@MONET ~/Documents/Research/Projects/Chromatin/Code (master)
$ git count-objects -v
count: 65
size: 68423
in-pack: 712
packs: 1
size-pack: 3040
prune-packable: 0
garbage: 0
Much better!
Posted in Software Development
Tagged chromatin, git, troubleshooting
Comments Off on git remove binary files
Friday 03/21/14
10:30a
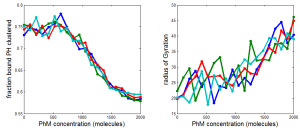
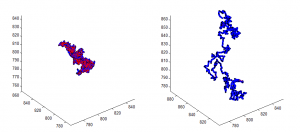
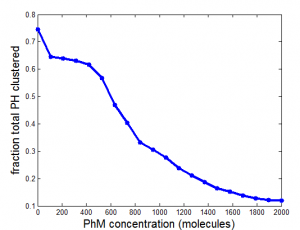
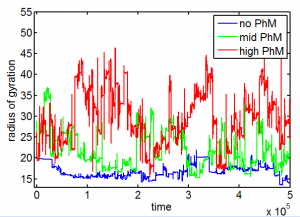
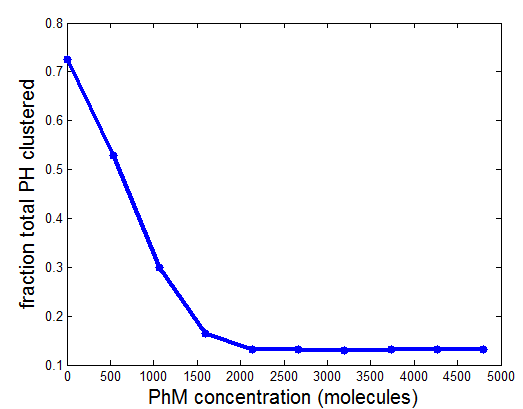
PH project
- results at greater sampling depth, longer simulation time, for the current published table parameters
- see PH project version corresponding to current date for details.
Data\2014-03-17_PhSim\PhCell_CappingModel5.mat
Research Organization
- Reorganizing projects folder
Freezer space organization
- started new box: Lib3
Notebook
- updated to notebook to wordpress jetpack to enable jetpack markdown.
- Previous plugin markdown on save discontinued to branch into jetpack markdown. We’ll see how this goes
Chromatin Project
- finish probe making. Concentrations look good.
Thursday 03/20/14
10:15a – 12:15a
Project 2
- see protected notes
Chromatin Project
Probe Making
RTs
- 4 uL primer
- gel looks good, basically complete incorporation. Maybe try 8 uL primer next time? (going to need to order more primer!)
- probes in 4C walkin, still need to do final clean up and concentrate with oligo-cleanup kit
- remaining 10 uL RNA in -20C. Should decide what to do with this or move it down to -80C tomorrow.
Data Analysis
- Finished analyzing F11-F12 2 color data (from Dec!).
- Completed ChromatinCropper’ analysis of F12-G01 data (more backgroundy but at least lots of localizations).
- still need to compile results and make some comparative figures
- still need to chose out the beauty shots from these rounds.
- might want to repeat next with the BXC related series in multi-color
Physicists of Biology hangout 03/20/14
Pallav Kosuri (Zhuang Lab)
Intro
- passive elacticity — stored energy in (in-elastic) muscle.
- contrast running to yoga.
- muscle structure z-bands spanned by titin. Titin stretches elastically.
Atomic Force Microscope and Titin
- Force clamp setup: fixed force (through feedback, measure force based on deflection of cantilever.
- Titin unfolds is fixed stochastic steps under constant force.
- remove the force protein collapses back to unstretched length in a single step. (entropic recoil, as opposed to instantaneous refolding).
- Titin made of Ig domains (very common structural motif, also found in antibodies).
- each unfolding step is a ~10 fold increase in length.
Mechanism for mechanical memory
- stems from oxidative stress (during exercise)
- mediated largely by glutathione (absorbs reactive oxygen species to protect cells) Oxidized gultathoine attaches to protein cysteines.
- most cysteines in titin are buried inside the folds. But are still gultathioni-ated during excercise stress.
- leave exposed to glutathione for a long time (40s instead of 2 seconds), domains don’t refold. (pull again and they instantly reach full extension).
- mutate the cysteines, remove this effect (happily refolds as well as it used to).
- cells have an enzyme GRX to remove glutathione from proteins — allows regulation.
test on heart cells
- step length, measure tension/force. Increase in force followed by relaxation.
- add glutathione muscle becomes much more pliant
- add DTT (remove all gulathionalition) become stiffer than usual.
Darren Yang (W. Wong Lab): Investigation of von Willebrand Factor in Hydrodynamic Flow
Intro: VWF
- multimeric blood glycoprotein, serves as a ligand for platelet adhesion and aggregation in vascular injury.
- Domain structure: A1 — binds to platelet cells. A3 domain binds to collagen. A2 domain – has a cleavage site.
- Hydrodynamic stress in blood flow. Shear flow (even volume), elongation flow (narrowing volume), tethered flow.
- molecule extends under velocity / shear flow.
- experimental setup — DNA stretches under elongational flow.
### vWF under hydro stress - when secreted, protein is cleaved by proteolytic enzyme. Cleavage requires protein unfolding.
- Q -> difference in A2 domain vs. rest of the protein to unfold under stress.
- built microscope compatible with conventional desktop centrifuge.
Rotem (Jeremy England lab)
Intro: how things move in cells
- standard model — thermally driven diffusion.
- dense environment, lots of motors, ATP hydrolysis, far from equilibrium
- Questions
- do proteins diffuse?
- do thermal fluctuations drive the motion?
- FRAP – motion of some proteins fits a diffusion model (e.g. Lippincott Schwatrz Science 2003). Better predicition for GFP than ‘functional proteins’
- recent single molecule tracking Parry et al Cell 2014. Treat cells with DNP / (no ATP) or even stationary growth phase, motion of protein becomes much more localized.
Approach:
- fluorescent photo-convertable proteins.
- take a collection of proteins fused to photo-convertable dendra (DDR). Study in ~7 different conditions.
observations
- HSP70-dendra propigates much slower than free dendra.
- fit to exponentional the decay rate of fluorescence in the photo-converted spot.
- diffusion rate not related to size of molecules.
- ATP depletion might lead to immobilization of a sub-population of dendra.
Sudha Kumari (Irvine MIT and Dustin Labs, NYU)
Background
- cell biologist interested in immune cells
- basis of specific recognition of ‘antigen’
- intra and inter-cellular signal amplification of antigen recognition
- physical contact? — a ’tissue’ lacking physical contact.
- Wiskott-Aldrich syndrome protein (WASP) mutant — T cells fail to amplify signal.
- WASP aids in polymerization of actin
- observe small patches of actin at Tcell contact interface in WT cells, lost in WASP mutants
- Actin patches correlate with TCR (signaling) micro-clusters.
- pattern antigen on the surface. (keeps clusters from moving / aggregating which was intefering with tracking).
- FRET polarization / anisotropy imaging in the patch. FRET pair have different alignment and different anisotropy. Patches have low anisotropy. Patches are ordered structures. Compartmental amplification?
- down regulation of WASP observed at time of aggregation of cells (cell-cell amplification part of the process).
- remove WASP, more cell-cell contact,
- less affinity to antigen presenting surface? (competition model)
- changes existing affinity between cells? (independent model)
- remove wasp, cells stick to eachother more (WASP prevents cell-cell sticking).
- mix WT cells and WASP negative cells, the null cells still stick to the positive cells.
Posted in Seminars
Comments Off on Physicists of Biology hangout 03/20/14
Protected: Project 2 controls 10/20/14
Posted in Genomics
Tagged project 2, troubleshooting
Comments Off on Protected: Project 2 controls 10/20/14
Wednesday 03/19/14
10:20a – 11:30p
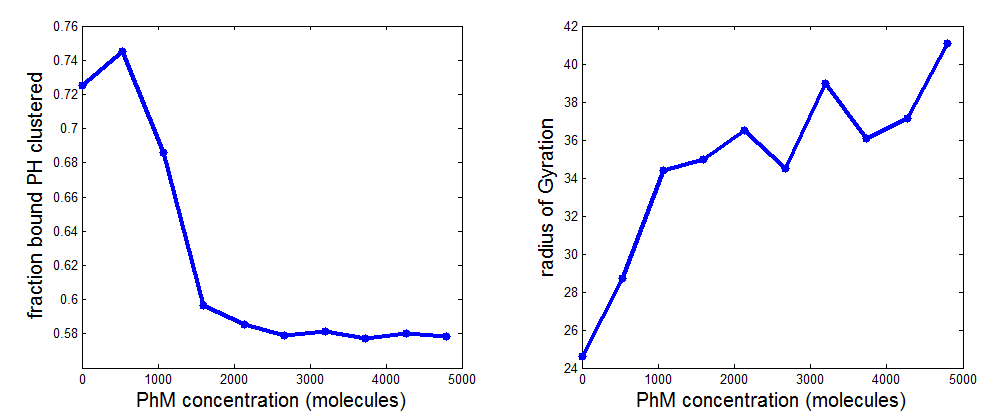
PH project
Simulations / model description
- Improved behavior from PH simulation
\Data\2014-03-17_PhSim\PhCell_CappingModel4
Main Figures
- wrote new scripts to graph the data in figures 1-3 quickly. Useful for changing binning intervals, fonts / colors / sizes etc easily.
- updated figures 1-3 to match new layout requests, fix match true-black, make scalebars equal width, match fontsizes, match panel spacing, correct cropping etc.
- sent new figures to Ajaz.
- BK currently reviewing manuscript.
Oligo-Secondaries
- working on figure
- wrote caption for current figure draft
- emailed draft and caption to Ting and Brian
Computer upgrades
- discussed windows server upgrades with CDW rep
- Ordered windows server updates
- need quote for server (can get remote desktop licenses through HHMI CDW portal).
- requested new quote with HHMI as bill to.
- configured remote desktop timeouts on cajal: http://technet.microsoft.com/en-us/library/cc754272.aspx
- disconnected state: 5 days until termination
- idle state: 2 days until disconnect
Chromatin Project
- set-up PCRs for probe making
- PCRs work (old get not so good and a bit overloaded). Some high MW product.
- also most samples mostly bubble band — expect 122 bp. Some samples have some low MW product, I wonder if this is mispriming? Should sequence this library too (MiSeq kit on the way).
- ran gel cleanup.
- set up T7 reactions to run O/N with RNasin inhibitor. Run started ~10:00p.
Data Analysis
- running bead-data compression for recently split multi-color data.
- Attempt resuming F12F11 data. Still missing 647 data for image 2. Now analyzing.
Posted in Summaries
Comments Off on Wednesday 03/19/14
Tuesday 03/18/14
9:30a – 6:30p, 11:00p – 1:00a
Oligo Secondaries
- work out new strategy for figure
- need simulations of binding density
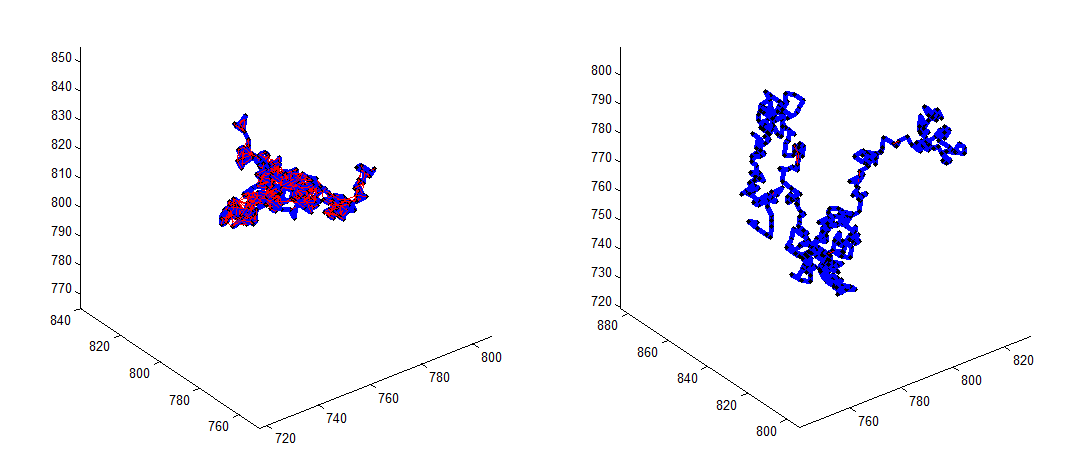
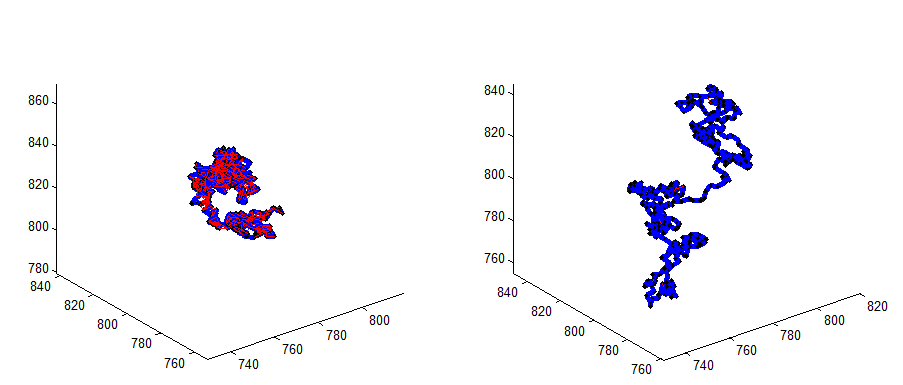
Chromatin Polymer simulations
Goal: illustrate effect of localization counts
* wrote new basic StickyPolymer simulation function
* wrote some new polymer plotting functions
* wrote a basic (stub) binding chemistry function
* all new PolymerSim package is in matlab-functions
Figure layout
- working on figure presentation
- see protected notes
Project 2
- see protected notes
Ph Project
- drop off still a little too shallow (previously this was always too steep).
- effect on chromatin structure was more obvious with lower bond energy (higher breaking probability).