11:00 am – 1:15 am
PcG RNAi
New Knockdowns and cell culture
- plate Ph, Pc, and mock RNAi cells onto coverglass
- density looks okay modulo a decent number aren’t sticking
- isolated cells for RNA extraction
- fixed plated cells to coverglass for imaging
- Psc null cells
- starting to dye for not changing media (despite growing too slowly to be confluent).
- replaced media on small plate and scraped plate (both these look pretty decent toward confluency).
- combined media into new small vial, maybe detached cells will attach and settle (took > 24 hrs to attach last time)
- made new Psc media (using modified Schneider’s from Lonzo and HI FBS).
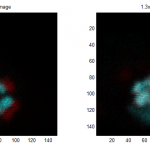
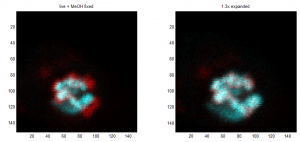
Imaging BX-C in Ph-KD
- finish imaging of L2F03F04
- start imaging of L4E24
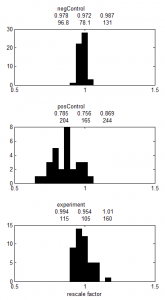
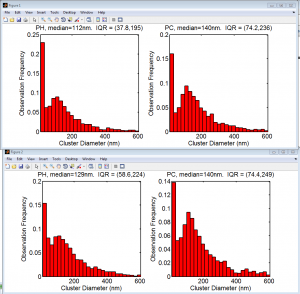
Analysis of BX-C internal domains in Ph-KD
- still fiddling with export options for ChromatinCropper2
- working on building ChromatinCropper2 GUI. This always takes longer than I think.
- meanwhile I’m working out what I hope is a better general system for GUIs.
Cell cycle studies
- prep cells for hybe
- hybe BX-C to cells.