9:00 am – ?
Getting Drosophila expression data
- Failed to find assembled FPKM data
- downloading 0-4hr data sets from modENCODE
- What’s with the big time window. There must be better these?
- need a GTF file for cufflinks to parse the SAM data
- running cufflinks: errors
- need to convert SAM to BAM
- downloaded GTF from UCSC refFlat.txt – this gives errors
- downloaded R5 data from ENSEMBL (release 5 is getting hard to find now everything defaults to release 6) ftp://ftp.ensembl.org/pub/release-77/gtf/drosophila_melanogaster
- RC Odyssey is very slow today. Was giving lots of acct errors too: server not responding. And my jobs stay in Pending purgatory for an hour. Better than all day I suppose.
Probe Design
- finished design of probes, check probe length
- still have hundreds with good length
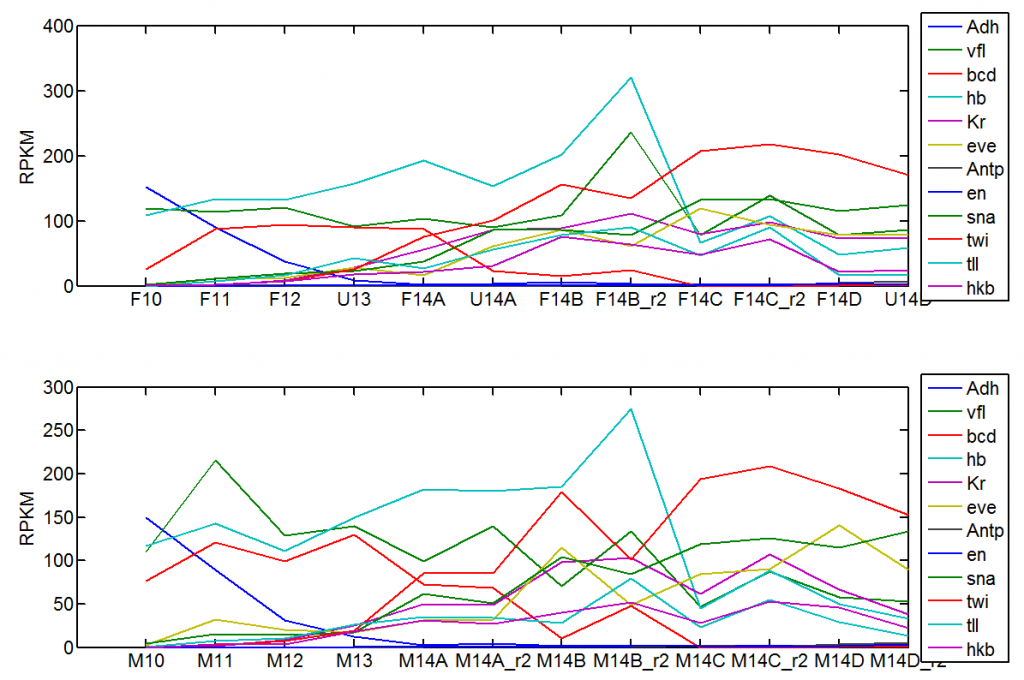
- computed sum FPKM from slicing data (this data is truly blastoderm — Hox genes and Segment polarity genes inv / en are cold off. Would be lovely to have some later embryos. Shame the modENCODE data is so broad. How about some single embryos at close timepoints?
- downloaded blastoderm single embryo data from Susan Lott’s paper
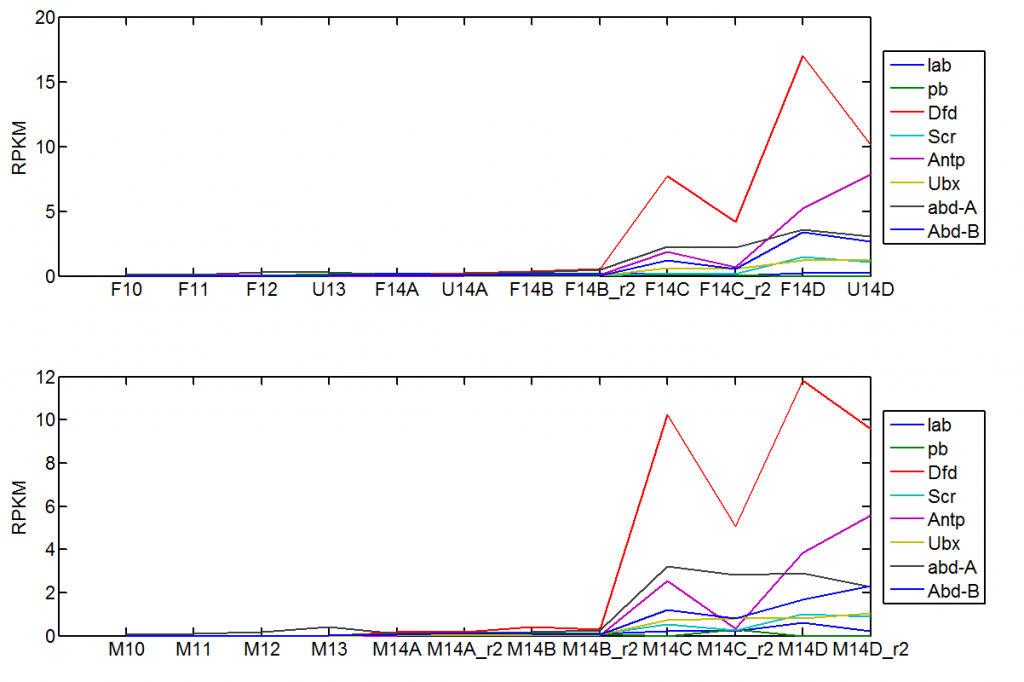
Hox Activation
Ordering control probes
- submitted orders to Alec for BioSearch / Stellaris probes for Kr and tll as positive controls.