September 2025 M T W T F S S « Aug 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 Categories
- AP patterning (13)
- Blog (1)
- Chromatin (88)
- Conference Notes (72)
- Fly Work (54)
- General STORM (25)
- Genomics (134)
- Journal Club (22)
- Lab Meeting (66)
- Microscopy (79)
- Notes (1)
- probe and plasmid building (58)
- Project Meeting (3)
- Protocols (13)
- Research Planning (74)
- Seminars (21)
- Shadow Enhancers (59)
- snail patterning (40)
- Software Development (5)
- Summaries (1,412)
- Teaching (9)
- Transcription Modeling (40)
- Uncategorized (10)
- Web development (19)
Links
Tags
analysis cell culture cell labeling chromatin cloning coding communication confocal data analysis embryo collection embryo labeling figures fly work genomics hb image analysis image processing images in situs Library2 literature making antibodies matlab-storm meetings modeling MP12 mRNA counting Ph planning presentation probe making project 2 project2 result results sectioning section staining shadow enhancers sna snail staining STORM STORM analysis troubleshooting writing-
GitHub Projects
Protected: some analysis of Hao Lib3 and 4
Posted in Genomics
Comments Off on Protected: some analysis of Hao Lib3 and 4
Friday 02/21/14
10:15a – 5:50p
Chromatin Project
Library 3 prep
- finish annotating new lib3 yellow green regions
- ran probe folding for yellow green regions
- Current probes (including 10 new black, 8 new blue): 91144
Deep Seq analysis
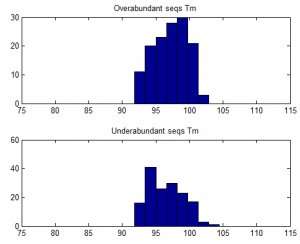
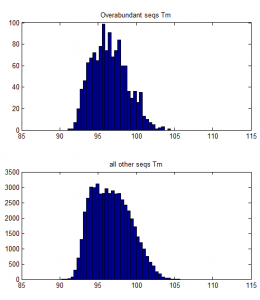
- Trying to understand why some sequences are consistently over-abundantly amplified.
- no effect of number of hairpins or number of dimers
- weak affect of Tm / %GC. Higher TM more GC more a bit more frequently abundant on ave.
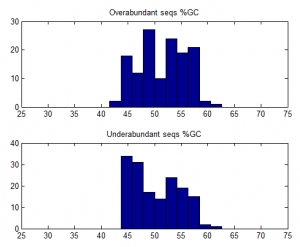
over-abundant after PCR vs all other seqs:
Analyzing data from Hao Lib
- see post
Data cleanup
- running splitdax on old bead data (this is the only way we can compress the bead movies).
- using D11 as test set.
Ph project
- spot checking of half a dozen or so of the S2 and WT Ph-STORM + FISH experiments don’t show obvious colocalization. The labeling is obviously sparser, (maybe 1/5 the number of clusters) probably just the brightest ones.
Thursday 02/21/14
5:30p – 8:00p, 9:00p – 10:30p
(snowshoeing).
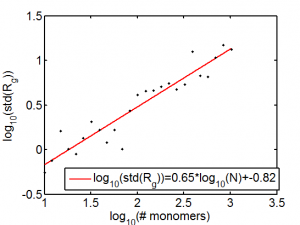
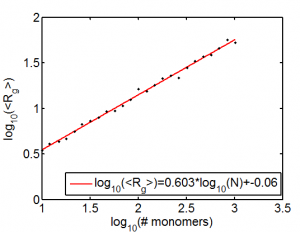
Random Walk Polymers
- Quick reference with the scaling of the self-avoiding RWP (exponent = 3/(2+d), where d is the spatial dimension). Thus 0.6 for 3D SAW polymer
- More interesting stuff http://arxiv.org/pdf/0711.3679v1.pdf
- Pivot + BFM models of random walks fit the appropriate scaling
Should do some more reading to double check if standard deviation of Rg scaling is right. Also could probably have used more runs to better determine this.
Wrote to Jane and student Baris about references for polymer models.
Wednesday 02/19/14
9:45a – 11:45p
Goals Today
- Final prep for meeting with Xioawei
- add slides showing density
STORM analysis
- launching drift bead analysis for F05 / F06 images and Ph-iFISH data
- launching split-dax for all un-analyzed datasets on RAID2 and Pro8.
- running z-calibration on new z-calibration data.
More Y regions
- chr2L:2210964-2237817 length: 26853
- chr2L:2489241-2499536 length: 10295
- chr2L:2967533-3000280 length: 32747
- chr2L:5517877-5549514 length: 31637 (Pc flank, big black)
- chr2L:8353587-8391384 length: 37797 (high gene, high express)
- chr2L:15254575-15274520 length: 19945 (isolated yellow in middle black);
- chr2L:13363559-13411277 length: 47718
- chr2L:18672197-18714928 length: 42731
- chr2R:4972056-5054981 length: 82925
- chr2R:9019018-9125165 length: 106147
- chr2R:13979742-14072371 length: 92629
- chr3L:9344016-9505324 length: 161308
- chrX:16161627-16351359 length: 189732
- chr2L:10203488-10443191 length: 239703 (big pretty solid yellow striped by green)
Feedback from XZ-meeting
- compare to more existing models — what scaling do they predict?
- see email to Bogdan
- see notes
- Discussion with Bogdan about project status (~8p – 10:30p)
Protected: Draft outline for chromatin paper
Posted in Chromatin
Tagged chromatin colors
Comments Off on Protected: Draft outline for chromatin paper
Tuesday 02/18/14
9:45a – 7:00p, 9:00p – 12:20a
STORM
- Imaging G123 (sample 1 from last staining). Very, very high nuclear background (and strong signal). surprising given the concentration used, but maybe should try 1/10th concentration.
STORM analysis
- Cajal crashed, all jobs got aborted.
- relaunching F05-F06 F06-F05 and S2 and WT data anlysis
- quick check of F06-F05 data, 750 localizations drop DRAMATICALLY AGAIN. Really the 750 buffer dies 50 fold faster.
Data analysis
- working on rough outline / scope of manuscript: see post
- working on figs for meeting with XZ (see google docs).
To do
- reply to JTB about review.
STORM Users planning meeting
STORM2 Max Laser Powers
- 488 laser head (100%): 580 mW
- 488 before expander: 275 mW
- 488 microscope backport: 180 mW
- 561 at head (2100mW in GUI): HIGH
- 561 laser at beam expander: 1500 mW
- 561 laser at backport 900 mW
- 647 laser at head (1700 mW GUI): HIGH
- 647 laser at beam expander: 720 mW.
- 647 laser at backport: 460 mW
- 750 laser head (2350 mA GUI): HIGH
- 750 laser at beam expander: 300 mW
- 750 laser at backport: 115 mW
TSTORM will go on STORM4
STORM2 will await a new functioning 750 setup before rebuild.
Posted in General STORM
Comments Off on STORM Users planning meeting
Monday 02/17/14
9:30a – 12:45p
Lab organization
- measure laser powers on STORM4
- STORM users meeting — see summary sent in email.
Ph project
Paper to do
- We need to make the sup. fig1. showing no pri-antibody controls.
- added the flag in S2 cell images
- don’t have all the no primaries with DAPI, need to repeat
- Color of both images in Fig1 should be same. May be we should just keep the color of anti-FLAG as well as anti-PH staining as red.
- will make everything red in future
- I still need to put number at couple of places for chip-seq data.
- Ajaz will handle
- Need to fix the ticks on y-axis in Fig2.
- will do in future
- Black lines of bars in some graphs of cluster diameter are not there, can you check that.
- fixed this in Fig 1.
- will check other graphs in future.
- Review latest draft of text
- done. Fully revised all sections / methods / captions. Sent to Ajaz.
- get better PcG-body images of Ph in S2 cells.
- done. some cells a little bit backgroundy in the cytoplasm but the nuclear enrichment is clear.
- some cells have body like structures
Data analysis
- Analyzing on Cajal:
- F05-F06 two color data 750 chn
- F05-F06 two color data 647 chn
- F06-F05 two color data 750 chn
- F06-F05 two color data 647 chn
- S2 PH-STORM FISH
- PH-WT FLAG STORM FISH
- running at 80% CPU on Cajal no stalling :).
Chromatin project discussions
- Discussion with Pallav about teaming up on chromatin
- Write to JK about yeast chromatin topology / interpreting the yeast HiC papers.
Saturday 02/15/14
5:50p – 11:30p
(Sailing day)
Deep Sequencing analysis
- continuing analysis of sequencing data, looking at F11 probes.
- Windows navigator refuses to browse to Data folder. Might have to do with the current read write operations from this folder. Still pathetic.
- Followed by blue screen of death computer crash. Go Windows!
- extrpolate probe names.
STORM
- 647zcal_0001 at imaging depth
- 647zcal_0002 at 800nm closer to surface
- 647zcal_0001 at imaging depth, replicate scan
- distance of imaging depth from beads, approx -1000 to -1200 nm
Posted in Summaries
Comments Off on Saturday 02/15/14