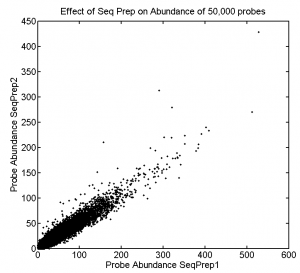
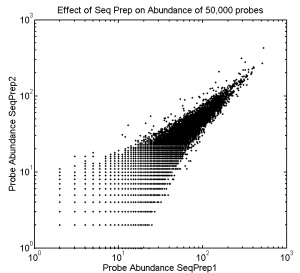
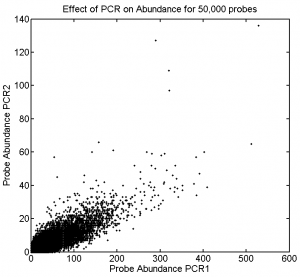
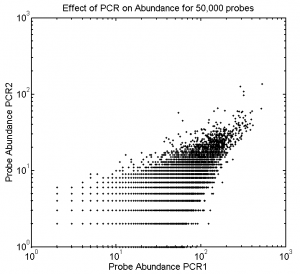
1:30p – 7:00p, 10:30p – 12:10a
Chromatin Project
Library3 Prep (renewed)
Revised gene list
Code updating
- Fixed minor bug in BatchLaunchOligoArray: ProcessTimeOut for blastall was in seconds but for java was in minutes.
- Fixed bugs in GetLocusSenseSeq — had extended to flip genes that are only partially contained within the locus, but hadn’t set the sequence start / end parser to max out correctly at each end.
- now running as BuildLib3_1401216 (should be 140216).
- some probe sets badly crash my computer. No idea why. Let’s try using the run in 1kb segment version of the code, I think that was generally more stable, and when crashed doesn’t lose the whole gene, just a bunch of regions. It will be more work to stitch these back together but I’m sure I can write a short script to do it.
STORM
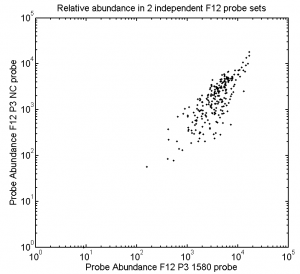
- Finish O/N STORM of F06-A647 F05-A750. 647 channel at least looks good through the end. see how these 750s came out.