September 2025 M T W T F S S « Aug 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 Categories
- AP patterning (13)
- Blog (1)
- Chromatin (88)
- Conference Notes (72)
- Fly Work (54)
- General STORM (25)
- Genomics (134)
- Journal Club (22)
- Lab Meeting (66)
- Microscopy (79)
- Notes (1)
- probe and plasmid building (58)
- Project Meeting (3)
- Protocols (13)
- Research Planning (74)
- Seminars (21)
- Shadow Enhancers (59)
- snail patterning (40)
- Software Development (5)
- Summaries (1,412)
- Teaching (9)
- Transcription Modeling (40)
- Uncategorized (10)
- Web development (19)
Links
Tags
analysis cell culture cell labeling chromatin cloning coding communication confocal data analysis embryo collection embryo labeling figures fly work genomics hb image analysis image processing images in situs Library2 literature making antibodies matlab-storm meetings modeling MP12 mRNA counting Ph planning presentation probe making project 2 project2 result results sectioning section staining shadow enhancers sna snail staining STORM STORM analysis troubleshooting writing-
GitHub Projects
Sunday 02/09/14
10:20a – 9:40p
Chromatin project
- finish analyzing new ANTC data
- Analyze all remaining complete yellow domains
- Analyze G01, G02, G03, G04, G01+G02
- Progress notes: see protected post
Saturday 02/08/14
10:30a – 4:45p, 10:00p-11:50p
Chromatin
Deep sequencing of libraries
- finish gel extraction aborted last night due to mini-centrifuge failure.
Data analysis
- Analyze new ANTP data
- issue: need to fix integrated drift correction, have some weird errors
- interpolation was not correct — need to convert bead frames to image frames.
- drift correction now working smoothly on integrated bead data
- issue 2: contrast averaging is too a bit too strong.
- Contrast is good but images are getting compressed into very low bit-depth relative to available dynamic range.
- maybe we should integrate over 60 frames and divide by 20 instead of 60? Lots of bead frames can in fact be added together without risking saturation.
Ph Project
Confocal Imaging of S2 cells
Prep
- make fresh Glox buffer solution
- let’s see if we can get this to work in chambered slides with Glox buffer.
Samples imaged
- Ph-Flag-WT
- Ph-Flag-M
- Endog. Ph in WT
- Endog. Ph in M
- Psc in WT
- Pc in WT
- Psc in M
- Pc in M
Observations
- Ph-Flag staining generally much weaker than others. Not sure why, seemed fine by STORM.
- With the usual contrast issues of confocal, there is some evidence for bodies in all samples.
- Most pronounced in Ph-FLAG-WT.
- Pc-WT vs mutant also potentional difference.
Fly work
- flipped stocks.
- All survived, some have dry food debris carry over but hopefully will recover.
Friday 02/07/14
10:00a – 11:30p
PH project
Meetings
- Finalize slides
- discussion with XZ
- Add effect on cluster count instead of effect on cluster size.
Updates
- working on figure for the effects on cluster count
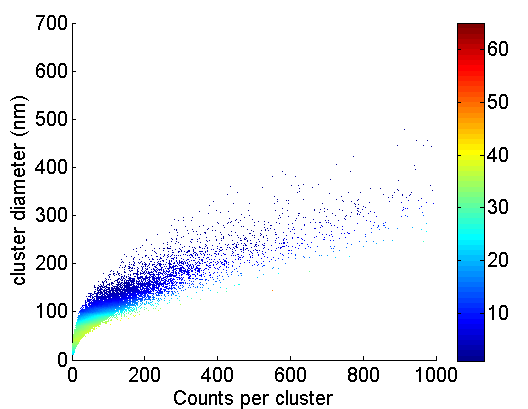
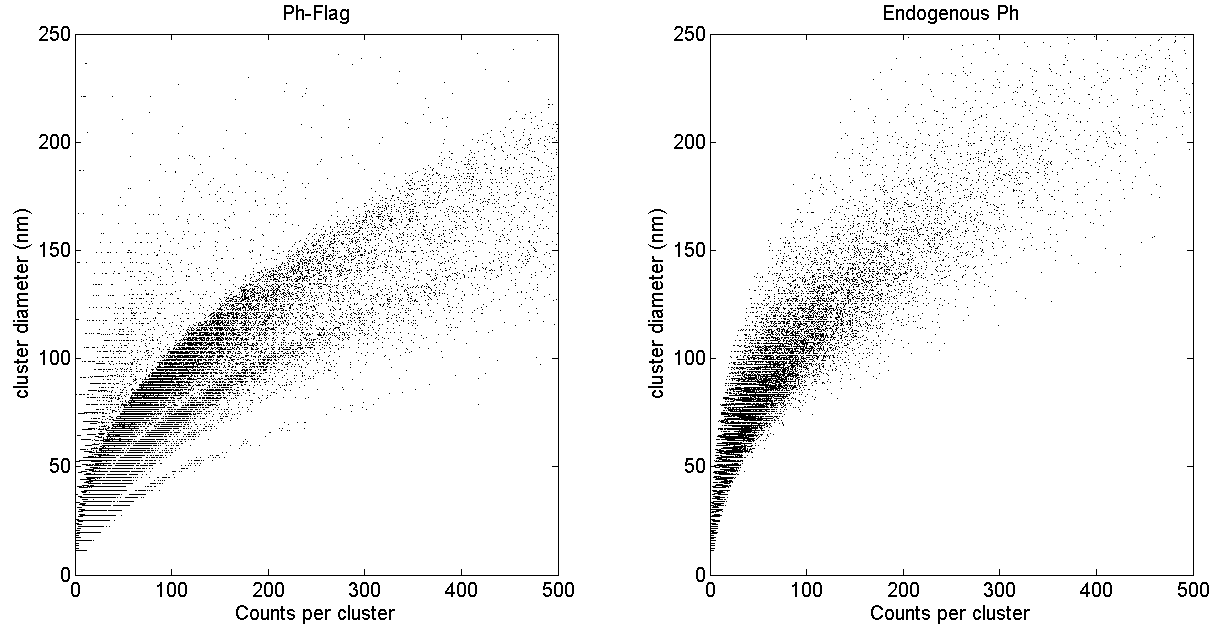
- added cluster counts vs. cluster diameter
- some further explorations in concentration effects:
Seminar
- Bodo Stern Seminar on getting published from the Editor’s perspective (former Chief Editor of Cell)
- see notes
Chromatin Project
Deep sequencing prep
- samples 1-8: A01-08 + B02-09
- samples 9-16: A05-12 + B03-B10
- 5 rounds of PCR
- ladder + samples 1-14
Final prep
- ran 2 more rounds of PCR for samples 1-8. (doesn’t seem to have made much difference?, probably could have done another 5, these were pretty dim).
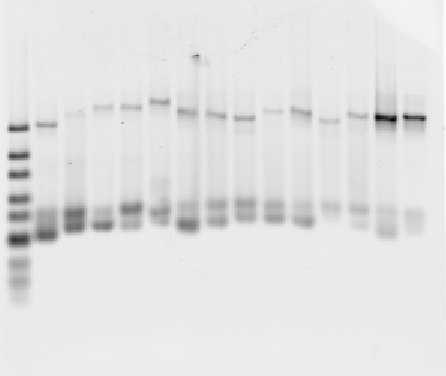
- Gel extracted all samples
- Centrifuge broke (all 3 lights flashing). Samples swallowed (~11:30pm)
- Next morning, (11:00a) extracted samples from centrifuge with emergency release at bottom (see manual), and finished eluting samples on alternative centrifuge. (hopefully these are still good. I guess we’ll know during the RT PCR)
Sample key
- Lib2-x1
- Lib2-x2
- Lib2-x1 v2.
- HaoLib4 E1
- HaoLib4 E3
- D12-P1
- G09-P1
- F11-P3 Nc
- Fll-P3 12-18
- F11-P3
- F12 P3 NC
- F12 P3 1580
- Hao L3-uni + L3 i1R
- Hao L3-uni + L3 i2R
- Hao L4 E1F + E1R
- Hao L4 E3F + E3R
Bodo Stern seminar 02/07/2014
Why Publish
- lay claims of priority (historic and present)
- responsibility to communicate results back to the public
- reach a broader audience than people in the same field
This talk
- not about tricks
- about editors and their task
- corresponding do’s and dont’s
Editor’s as judges
- Editor makes decision. Reviewers are purely advisors
- Editor’s are “not academics”,
Editors goals
- how significant is this advance
- How interesting is the paper across the fields — not just a quality of science judgement
Tips to get the message
- Take adventage of the cover letter!
- not seen by reviewers: can be broader, can give a punchline that is not being criticized by scientists for accuracy
- what was known before (more colloquial)
- not bad to exclude reviewers (generally honored)
- presubmission inquery
- can send to several journals at once
- be prepared to accept their advice if you ask for it.
- very often editor response will not be informative.
- Help the editor understand the logic
- some non-expert should be able to read and understand the paper.
- don’t just give it to people in the lab
- Make the figures self-explanatory and easy to understand
- editors have ways to review quickly. Often look at figures before any of the text.
- biggest problem with papers is the logic.
Editors as Mediators
- Editors would rather be on the side of publishing something that’s incomplete if it will be cited.
- Reviewers will always point out where the story could go.
- Editor’s need to take risks, just because it’s published in cell doesn’t mean it’s wrong’.
Editorial letters: invitation to resubmit
- Look for we would be happy to consider or we would be prepared to consider. Anything else is a rejection, regardless of positive feedback.
- if you want to go further you better get in touch with the editor.
- If positive, get in touch with an editor early
- be positive in line-by-line responses. don’t be aggressive, be constructive.
- email first, can set up a time to call and adress
Rejections
- if rejected: don’t respond within 24hrs. this will not go over well
- stick purely to science. Can present facts for why reviewer is incorrect.
- Take-home message: Be Assertive. Easier to be assertive if you know the editors (this is an advantage of senior faculty).
Meet and talk to editors
- Editor’s like to meet people in science. More editors are going to conferences.
- Editor’s will recruit papers that they really want
- certainly want new reviewers.
- review for journals you want to publish in.
- Journals have pet projects they want to publish more in. Meet editors find out what they are looking for.
- Bodo Cell didn’t use to publish a lot of single molecule. Less competition at first.
- new interests of journals not advertised on web
- get a sense of the editor and their logic as a scientist. See how you relate. Find an editor you respect.
On reviewing papers
- easier to find the flaws and plus than make decision.
- Goals:
- review the technical quality (this the editor wont overrule)
- comment on significance / context / clarity (these are also up to editor)
- don’t give a laundry list. At least sub-class them: critical, supporting, etc.
- don’t keep all the signficance etc arguments in confidential comments. Keep it all in the open review if possible.
- Ask people in your lab to help you and acknowledge in your comments to the editor who helped you.
- either keep confidential or sign it.
After publication. Online presence of scientific work
- Work with Harvard communications office (Peter Reuell preuell@fas.harvard.edu)
- good to get a press release.
- Add ‘final author version’ (i.e. your format not their layout / copy-editing). You always own this copy.
- Add paper to PubMedCentral (required by NIH, need PubMedCentral ID#s). all program officers will check that the papers are there.
Copyright and open access
- Harvard keeps copyright to everything (since 2008)
- Nature, Science, Cell require Harvard to waive this copyright
- NIH requires final-author version on PubMedCentral within a year (many journals do this automatically, but you need to check).
- number of scientists has tripled in recent years, Cell, Nature, Science have flat rates
Posted in Seminars
Comments Off on Bodo Stern seminar 02/07/2014
Thursday 02/06/2014
10:00a – 12:50a
Ph Project
- Meeting with Ajaz, discuss paper revisions.
- continue preparing slides for discussion with XZ.
Meeting notes (4p – 7:30p)
To image by Confocal (top priority)
- Endogenous Ph
- Endogenous Pc
- Endogenous Psc (may not work)
- Ph-Flag
- Ph-M-Flag
figure tweaks
- increase contrast in Fig 1 F.
- Thicker scale bars on all panels in Fig 2
- Put more heavily dispersed Ph into Fig 5.
- Fig2 Panel I find example with stronger colocalize
- better Y axis label for histograms.
controls for supplement
- Supplement figure S1.
- No primary controls
- Flag in S2 cells with DAPI contrast.
Fig 5
- change figure back to Ajaz format
- Refer to supplement.
- don’t want to do review experiments to show chromatin template effects
Model for supplement
- Chromatin template important or not?
- Effect of clustering on radius gyration
Expression analysis
- Ajaz will send raw expression files.
- see if we take top 30 Pc target genes, how many increase expression vs. the average gene. Something like this.
Chromatin Project
Sequencing Prep
- Get library from Hao
- check primer concentrations
- 2x Plate PCR of library. D01 – G12
- ran 25 uL scale PCR with T7 and common primers
- pool these, add on T7-NEB adapter in 3 rounds of PCR
Lib2 probes to sequence
- F11-P3 3 replicates of probe synthesis
- F12-P3 2 replicates of probe synthesis
- G09-P1 1 probe sample
- D12-P1 1 probe sample (top left of orange box)
- all diluted 0.5 uL to 200 uL ddH2O
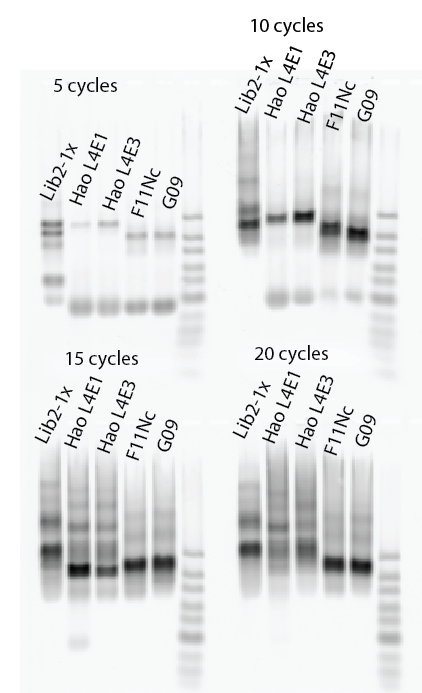
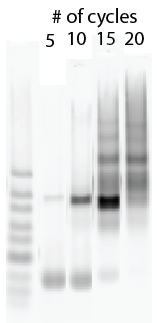
Poor man’s limited cycle PCR:
- Lib2X, Lib4E1, Lib4E3, F11NC, G09
Focus: birth and death of a probe in PCR
LC-PCR
- Lib2-x1 1:32, (2 uL)
- Lib2-x2 1:32,
- Lib2-x1 1:32 v2. (2 uL)
- HaoLib4 E1 (8 uL)
- HaoLib4 E3
- D12-P1 (2 uL)
- G09-P1
- F11-P3 Nc
- Fll-P3 12-18
- F11-P3
- F12 P3 NC
- F12 P3 1580
- Hao L3-uni + L3 i1R
- Hao L3-uni + L3 i2R
- Hao L4 E1F + E1R
- Hao L4 E3F + E3R
Completed limited cycle PCRs and gel extraction and cleanup.
Creating effective slides – Jean-Luc Doumont
Intro
slides from googling harvard
- examples (mostly with too much text)
On bullets
- looks like a list, it’s not a list
- don’t let ppt drive the way you present information
- multiple topics — 5 or less. (can’t count more than this in a spot)
- make a hierarchy if you have move.
- doesn’t promote splitting into separate slides
Details / asides
- contrast / format
- prefer sans serif (feet) (serif has variable width, not good for contrast).
- no funny backgrounds, new format bullets.
- no vertical text
A simple test
- if I remove the slide, can you redraw what was showing?
Creating effective slides
Foundations of communication
- Goal: get the audience to understand a message
- pay attention (presentation it’s now or never)
- be able to act upon your content (ask questions, start collaboration, talk to you etc).
- message is the interpretation about the information.
- not the “what”
- focus on the “so what”
- is this ‘good news’ is this ‘difficult, impressive?’
- on graphs
- disconnected legends not good. (put the labels next to the objects)
- what do you conclude from this graph?
- maximize message for audience
- strict selection.
Three rules
- Adapt to your audience
- aside: story sending photos me.jpg. be audienced focused
- Noise
- speaker’s way of distracting the audience
- repetitive gestures
- filler words
- excess movement on slides
- laser pointers (no laser’s for me)
- increase the signal.
- are the boxes necessary?
- inconsitencies on slide — (time spent not on content)
- parantheticals. Not necessary
- Effective redundancy
- compensate for zone out loss
- Balancing text and slides
- get message just from slide
- get message just from audio.
- periodically blind people (taking notes)
- if it’s too small to read 6 a page, it’s too small
- ask a friend if the ‘what’ is clear, and if the ‘so what’ is clear.
- practice at least once without the slides. Otherwise you are not a speaker. You are a museum guide.
- (Make people curious about the slide before showing it).
- Do you know what your next slide is? without looking at laptop, good eye contact.
Common problems
- slides for speaker instead of audience. (cross out things you negate)
- slides that double up as the written report (big problem in the business world).
- copy paste from elsewhere
- take time to do them well or don’t do them at all.
- mismatch slides from previous presentations when in a hurry.
- better approach: no slides.
- First who is coming etc.
- Fill in blanks about content (1 sheet of paper) what is the plan
- If this is all you have, practice. –> How about matching expectations?
Presentation structure
- Higherarchy: main message, main points
- More time, don’t add more slides, add more points.
- where do you put the so what? = title. Audiences read titles.
- title is not redundant with the slide info
- state the so what. A recent alarming drop. Oh yes, I can see that in the graph.
- state, then develop. Not at the bottom.
- don’t squeeze the title into the top, don’t segregate it away with the line.
- LEFT align title easier. Complete sentence. Two lines okay (3 not). Good line breaks.
Designing the slides
- common mistakes:
- Noisy template
- dump ideas randomly into slide.
- what is a template?
- A set of rules, where the content goes, and how it’s formatted.
- if there is no content, there is nothing on it.
- Title of slides first (what are you trying to tell the audience).
- where do we put the data?
- big stuff align with title.
- small stuff has only 1 alternative, further in.
- more on graphs
- add tick marks only as needed.
- move axes if necessary (now it goes to 100% cause it hits that number).
- annotate graph, it says something about the data.
- This is visual, this is how we do it on the chalkboard. ‘a slide is more like a chalk board’.
Using the slides
- Timing
- Pointers? (do clear slides need pointers?)
- laser pointers run around
- Self / stance
- eyes don’t come back to you. They stay on the slide.
- eye contact is the major connection of credibility. Stable, tall, look in the eyes.
- way to convince audience in a talk is not your data, it’s the eye contact.
- stand square to audience. Point with hand, point with your eyes. When your eyes go back to the audience, they track back to you.
- don’t look at audience when standing infront of side. They aren’t supposed to be looking at you at this point. or reading the text on your face.
- even sticks return to you. Though have a place where you put it down!
Three things to check
- Does it convey a so what (ideally a full sentence in title. up to two lines).
- Is it developed visually?
- Did we remove all the noise?
Questions
- backgrounds — dark backgrounds for dark rooms (fluorescence images), light backgrounds for light backgrounds
- white background generally best, keep a bright room, have good eye contact?
- what about switching? (best if you switch lights). People will be disoriented for a little bit.
- posters? use the format.
- get rid of everything people won’t read: e.g. references.
- if you really need it have a handout. – name email website, refs etc.
- readability should be readable on a US letter sheet.
- talk outlines?
- okay but please don’t make it boring.
- make it specific to your talk.
- don’t do it too early.
- the problem statement, the task, and then the takehome message
- it’s just for the stuff in the middle
- outlines help you summarize — it’s important to say it.
- outline for 6 minute presentation, of course, just not necessarily a slide. But announce the logic of your presentation (like mapping in writing).
- acknowledgements — problem, you’re audience doesn’t usually care about these.
- leave it up during questions and answers. Acknowedge when relevant.
- coauthors, present at beginning, say what each author did. That’s not the same as acknowedgments.
Posted in Seminars
Comments Off on Creating effective slides – Jean-Luc Doumont
Tuesday 02/04/2014
11:00a – 8:00p, 9:30p – 10:30p
Ph project
Manuscript writing
- working on model section
- finished description of first model (on-rate or off-rate limited)
- finished writing description of second model, ~3:30p, (chromatin availability limited)
- working on methods section
- wrote description of STORM imaging
- wrote description of antibody labeling
- wrote description of image analysis and clustering
- wrote description of modeling / polymer-simulations
- working on discussion
- Basic discussion of clusters needs overhaul.
Figures
- TO DO: Redesigning Figure 5 (model figure).
- change kinetic panels to first set.
- remove images (too many panels)
- redo plots in second set of models
Literature
- finished reading Tijan Lab Elife article on fastFISH.
- Nice kinetic measurements of RNAP binding, RNA appearance, RNAP release and RNA release.
- All with T7 on glass substrate, would be nice to eukaryotic polymerases (in vivo?) Probably requires a lot more signal and doesn’t require the ultra fast hybridization since T7 is blazing fast (binding 1 per second, elongating 300 bp/s or something like that)
- Pivot algorithm improvements
- Beautiful review on regulation of gene expression in the genomic context by Taylor Atkinson and Marc Halfon: http://journals.sfu.ca/rncsb/index.php/csbj/article/view/csbj.201401001
- Simon and Kingston: Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put.
- ‘central mechanistic question, how PcG complexes shut down transcription, has been a tough nut to crack’. (i.e. not yet worked out?)
- long standing idea of altered template accessibility called into question (no new citations here though, I suspect they follow in a more detailed discussion after the intro).
- PcG targeting in mammals more RNA directed (e.g. Xist, HOTAIR). Though deletion of HOTAIR + flanking regions has no phenotype.
- no direct evidence yet, merely correlative (knock this down, change H3K27me3). Want to see, mutate this binding site which affects interaction, change repression. Observe lncRNA directly at the site where the change (PcG binding) is happening.
- emerging evidence challenges the central role of K27me3 for targeting PRC1.
- Yuan et al 2012 study implies K27me3 accumulates after onset of silencing (sure, but PcG dependent silencing?)
- perturb K3K27me3 levels not dramatically disturb immunostaining of PRC1 (presumably in polytene).
- Pc and Ph unnecessary of ubiquitination activity of PRC1, and they compete for a position in the complex with KDM2, an enhancer of ubiquitination. (genetically though they are key elements of PcG silencing).
(8:00p-9:30p. Replace broken bike chain. Attempt and failed to replace worn out rear gear cassette)
Chromatin Project
Imaging
- STORM2 available again
- Finish imaging current recently stained cells (G05 and G09).
- Check age of fixed cells, repeat attempt of double stains
- Double stains done so far (none of which I’ve found time to analyze because of the Ph project take-over).
–
Monday 02/03/2014
10:00a – 11:50a
Seminars
- Hao practice talk for lab meeting, 11a – 1p.
- Tamara’s seminar at the stat depart colloquium, (see notes)
Ph Project
manuscript work
- working on re-arranging figure panels and colors
- added whole cell images to figure 2
- added more zoom-ins to figure 2
- introduced color conventions — each protein has a unique color. General color map is red to cyan.
- started working on rewriting modeling section text.
- Should be written from ground up
- Need to make it clear from topic sentence what the objective to test is (Ajaz has a good start for this I think), and then clearly state the two specific alternative hypothesis we will test through the model and its comparison to our data.
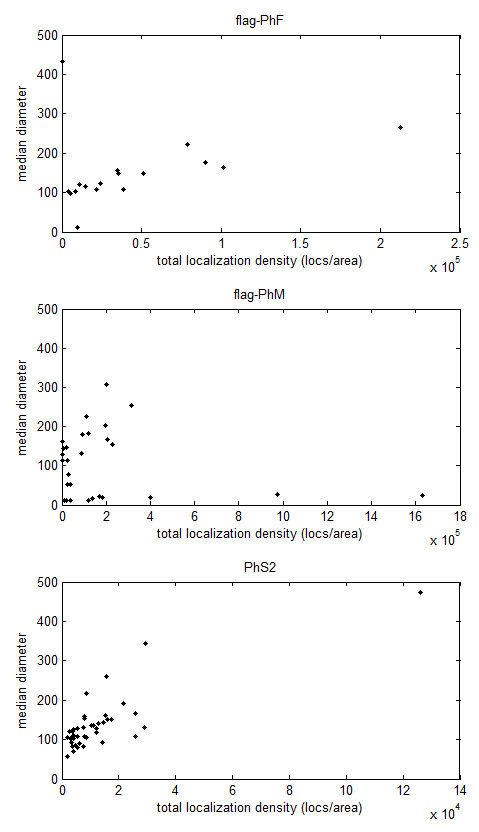
- Probably need to redo ‘fraction clustered’ in terms of new cluster diameter framework.
data collection
- continue imaging WT-Ph-flag + Psc
- take bead images
- start imaging M-Ph-flag + Psc
- Dave not talking properly to hal? STORM2 dave generated run file says to record 25,000 frames of 750. I come back two hours later and it’s still recording frame like 2 million+ of the first 750 movie. Not inspiring.
- Restarted imaging run. Seems to be working smoothly.
Chromatin project
STORM Imaging
- finish imaging ANTC. Lost focus at position 3 last night. :(. Reset focus and increased IR laser strength for focuslock.
- focus successfully maintained for a few more images (10a – 1p).
- realign camera and microscope. Camera removed today for use on STORM4-dual objective.
- image G01 G02 data on STORM4. Very good staining, very good switching. imaging 90K frames (could probably do more, but that’s most of it at least). Little 405 necessary.
- ISSUES with STORM4: GUI doesn’t actually update when switching parameters (for example, if the run shutters box defaults to be checked on a new parameter set, but not the current one, when you switch to the new parameter set the box will be unchecked. Nonetheless, it will still run as if it is checked. This problem is STORM4 specific.
- More annoying: I can’t set the conventional images and STORM images to have different movie styles. I set one to
.daxand one to.647and it sets them both either to whichever I’ve set more recently. This problem is also STORM4 specific. The only code I know of that’s different is the default parameters are set to point to the STORM4 defaults.
Cell culture
- passage cells
- (need to flip fly stocks very soon, tomorrow if manageable).